|
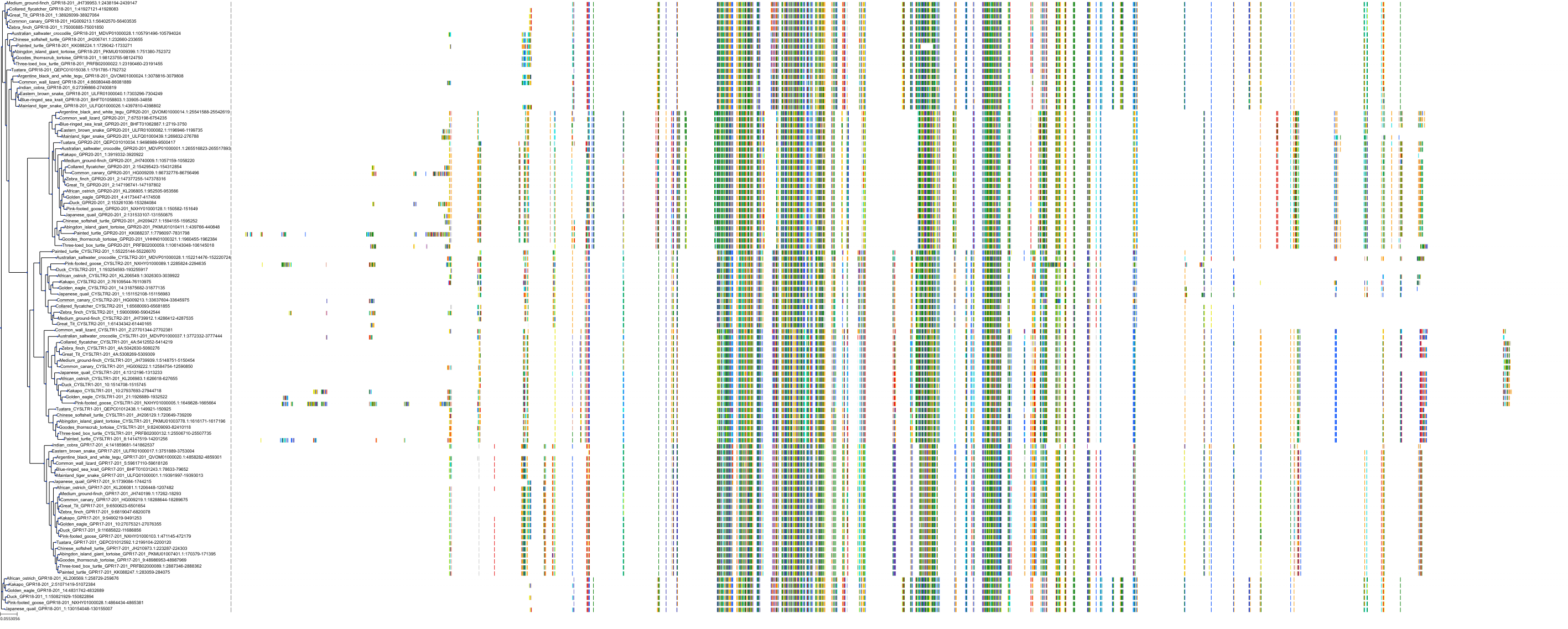
In vitro binding of cysLT1 receptor to guinea pig lung membranes.
|
Cavia porcellus
|
2.0
nM
|
|
|
Binding affinity towards Cysteinyl leukotriene D4 receptor (cysLT1) was measured by the displacement of [3H]LTD4 radioligand
|
Cavia porcellus
|
0.3
nM
|
|
|
Compound was evaluated for its ability to displace [3H]LTD4 from Cysteinyl leukotriene D4 receptor in guinea pig lung membranes
|
Cavia porcellus
|
0.3
nM
|
|
|
Displacement of [3H]LTD4 on guinea pig lung parenchymal membranes
|
Cavia porcellus
|
0.3
nM
|
|
|
Binding affinity against Cysteinyl leukotriene D4 receptor
|
None
|
0.3
nM
|
|
|
Antagonism of Cysteinyl leukotriene receptor 1 from guinea pig lung membranes
|
Cavia porcellus
|
2.0
nM
|
|
|
Inhibitory contractile effect of compound in guinea-pig tracheal strips induced by LTD4 (100 nM) (% inhibition obtained by comparison with control group)
|
Cavia porcellus
|
58.7
%
|
|
|
50% inhibitory concentration on the LTD4 (100 nM) induced contraction of guinea-pig tracheal strip bioassay
|
Cavia porcellus
|
1.03
nM
|
|
|
Compound was tested for its ability to block antigen induced airway obstruction in guinea pig at dose 1 mg/kg at 2 hr
|
Cavia porcellus
|
51.6
%
|
|
|
In vitro effect of compound on leukotriene-D4 LTD4 (100 nM) induced contraction of isolated guinea pig tracheal strip
|
Cavia porcellus
|
1.03
nM
|
|
|
Compound was evaluated for LTD4 induced guinea pig trachea contraction
|
Cavia porcellus
|
0.3162
nM
|
|
|
Ability to antagonize LTD4 receptors isolated from guinea pig lung membranes
|
Cavia porcellus
|
2.0
nM
|
|
|
Binding affinity against leukotriene D4 receptor in [3H]LTD4 binding assay
|
Cavia porcellus
|
2.0
nM
|
|
|
Concentration required for inhibition of binding of [3H]LTD4 to guinea pig lung membranes
|
Cavia porcellus
|
0.44
nM
|
|
|
Displacement of [3H]leukotriene D4 (LTD4) from receptor in guinea pig lung membranes
|
Cavia porcellus
|
2.3
nM
|
|
|
In vitro for antagonistic activity against LTD4 receptor in guinea pig ileum
|
Cavia porcellus
|
5.0
nM
|
|
|
In vitro inhibition of [3H]LTD4 binding to guinea pig lung membrane
|
Cavia porcellus
|
44.0
nM
|
|
|
Inhibition constant for displacement of [3H]LTD4 on guinea pig lung parenchymal membranes
|
Cavia porcellus
|
0.34
nM
|
|
|
Ability to mobilize Ca+2 in LTD4 dependent Ca+2 mobilization assay in differentiated human monocytic U-937 cells
|
Homo sapiens
|
1.0
nM
|
|
|
In vitro blocking of extracellular calcium mobilization in human U937 cells
|
Homo sapiens
|
1.0
nM
|
|
|
Compound was evaluated for influx of calcium mobilization in human U937 cells
|
Homo sapiens
|
1.0
nM
|
|
|
Compound was tested for its ability to inhibit calcium influx in human U937 cells
|
Homo sapiens
|
1.0
nM
|
|
|
PUBCHEM_BIOASSAY: Luminescence Microorganism-Based Dose Confirmation HTS to Identify Compounds Cytotoxic to SK(-)GAS Group A Streptococcus. (Class of assay: confirmatory) [Related pubchem assays: 1677 (Project Summary), 1662 (Primary HTS)]
|
Streptococcus
|
936.0
nM
|
|
|
DRUGMATRIX: Cysteinyl leukotriene receptor 1 radioligand binding (ligand: [3H]LTD4)
|
None
|
1.731
nM
|
|
DRUGMATRIX: Cysteinyl leukotriene receptor 1 radioligand binding (ligand: [3H]LTD4)
|
None
|
0.865
nM
|
|
|
DRUGMATRIX: Protein Serine/Threonine Kinase, ERK2 enzyme inhibition (substrate: Myelin Basic Protein)
|
Escherichia coli
|
538.0
nM
|
|
|
DRUGMATRIX: Protein Serine/Threonine Kinase, p38alpha enzyme inhibition (substrate: Myelin Basic Protein)
|
Escherichia coli
|
353.0
nM
|
|
|
DRUGMATRIX: Adenosine A3 radioligand binding (ligand: AB-MECA)
|
None
|
770.0
nM
|
|
|
Inhibition of human MATE1-mediated ASP+ uptake expressed in HEK293 cells at 20 uM after 1.5 mins by fluorescence assay
|
Homo sapiens
|
52.0
%
|
|
|
Competitive binding affinity to human PXR LBD (111 to 434) by TR-FRET assay
|
Homo sapiens
|
710.0
nM
|
|
|
Time dependent inhibition of CYP1A2 (unknown origin) at 100 uM by LC/MS system
|
Homo sapiens
|
11.0
%
|
|
|
Time dependent inhibition of CYP2C19 in human liver microsomes at 100 uM by LC/MS system
|
Homo sapiens
|
10.0
%
|
|
|
Time dependent inhibition of CYP2D6 (unknown origin) at 100 uM by LC/MS system
|
Homo sapiens
|
10.0
%
|
|
|
Time dependent inhibition of CYP3A4 (unknown origin) at 10 uM by LC/MS system
|
Homo sapiens
|
43.0
%
|
|
|
Time dependent inhibition of CYP2C8 (unknown origin) at 3 uM by LC/MS system
|
Homo sapiens
|
10.0
%
|
|
|
Time dependent inhibition of CYP2C9 (unknown origin) at 10 uM by LC/MS system
|
Homo sapiens
|
10.0
%
|
|
|
Time dependent inhibition of CYP2B6 (unknown origin) at 10 uM by LC/MS system
|
Homo sapiens
|
10.0
%
|
|
|
Antagonist activity at human CysLT1
|
Homo sapiens
|
1.9
nM
|
|
|
Antagonist activity against CysLT1 receptor (unknown origin) expressed in HEK293 cells assessed as inhibition of LTD4-induced calcium mobilization pre-incubated for 10 mins before LTD4 addition by Fluo-4 AM dye based fluorimetric assay
|
Homo sapiens
|
14.0
nM
|
|
|
Induction of mitochondrial dysfunction in Sprague-Dawley rat liver mitochondria assessed as inhibition of mitochondrial respiration per mg mitochondrial protein measured for 20 mins by A65N-1 oxygen probe based fluorescence assay
|
Rattus norvegicus
|
66.2
nM
|
|
|
Displacement of [3H]LTD4 from cysteinyl leukotriene receptor 1 in Hartley guinea pig parenchymal membrane after 30 mins by liquid scintillation counting method
|
Cavia porcellus
|
0.3
nM
|
|
|
Inhibition of cysteinyl leukotriene receptor 1 (unknown origin) expressed in HEK293 cell membranes after 45 mins by scintillation spectrometry
|
Homo sapiens
|
1.8
nM
|
|
|
Antagonist potency at human CysLT1R expressed in African green monkey COS7 cells assessed as inhibition of LTD4-induced Ca2+ levels after 60 mins by fura-2/AM-fluorescence based spectrofluorimetry
|
Homo sapiens
|
0.003981
nM
|
|
|
Antibacterial activity against Staphylococcus aureus MRSA ATCC 43300 (CO-ADD:GP_020); MIC in CAMBH media, using NBS plates, by OD(600)
|
Staphylococcus aureus subsp. aureus
|
36.84
%
|
|
|
Antibacterial activity against Escherichia coli ATCC 25922 (CO-ADD:GN_001); MIC in CAMBH media using NBS plates, by OD(600)
|
Escherichia coli
|
4.93
%
|
|
|
Antibacterial activity against Klebsiella pneumoniae MDR ATCC 70063 (CO-ADD:GN_003); MIC in CAMBH media using NBS plates, by OD(600)
|
Klebsiella pneumoniae
|
3.3
%
|
|
|
Antibacterial activity against Pseudomonas aeruginosa ATCC 27853 (CO-ADD:GN_042); MIC in CAMBH media using NBS plates, by OD(600)
|
Pseudomonas aeruginosa
|
41.4
%
|
|
|
Antibacterial activity against Acinetobacter baumannii ATCC 19606 (CO-ADD:GN_034); MIC in CAMBH media using NBS plates, by OD600
|
Acinetobacter baumannii
|
20.95
%
|
|
|
Antifungal activity against Candida albicans ATCC 90028 (CO-ADD:FG_001); MIC in YNB media using NBS plates, by OD630
|
Candida albicans
|
0.8
%
|
|
|
Antifungal activity against Cryptococcus neoformans H99 ATCC 208821 (CO-ADD:FG_002); MIC in YNB media using NBS plates, by Resazurin OD(600-570)
|
Cryptococcus neoformans
|
1.96
%
|
|
|
Antiviral activity determined as inhibition of SARS-CoV-2 induced cytotoxicity of Caco-2 cells at 10 uM after 48 hours by high content imaging
|
Homo sapiens
|
-3.07
%
|
|
|
SARS-CoV-2 3CL-Pro protease inhibition percentage at 20µM by FRET kind of response from peptide substrate
|
Severe acute respiratory syndrome coronavirus 2
|
-6.559
%
|
|
|
Antiviral activity determined as inhibition of SARS-CoV-2 induced cytotoxicity of VERO-6 cells at 10 uM after 48 hours exposure to 0.01 MOI SARS CoV-2 virus by high content imaging
|
Chlorocebus sabaeus
|
-0.2
%
|
|
Antiviral activity determined as inhibition of SARS-CoV-2 induced cytotoxicity of VERO-6 cells at 10 uM after 48 hours exposure to 0.01 MOI SARS CoV-2 virus by high content imaging
|
Chlorocebus sabaeus
|
-0.2
%
|
|