| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| UNII | W1B375O5M2 |
| EPA CompTox | DTXSID00179866 |
Structure
| InChI Key | HXHAJRMTJXHJJZ-UHFFFAOYSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C20H24BrF2N5O3S |
| Molecular Weight | 532.41 |
| AlogP | 3.86 |
| Hydrogen Bond Acceptor | 6.0 |
| Hydrogen Bond Donor | 3.0 |
| Number of Rotational Bond | 10.0 |
| Polar Surface Area | 109.58 |
| Molecular species | BASE |
| Aromatic Rings | 2.0 |
| Heavy Atoms | 32.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Vascular endothelial growth factor receptor 2 inhibitor | INHIBITOR | PubMed PubMed |
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Enzyme
Kinase
Protein Kinase
TK protein kinase group
Tyrosine protein kinase DDR family
|
- | 24-450 | 422 | - | - | |
|
Enzyme
Kinase
Protein Kinase
TK protein kinase group
Tyrosine protein kinase Tie family
|
- | 18-620 | - | - | - | |
|
Enzyme
Kinase
Protein Kinase
TK protein kinase group
Tyrosine protein kinase VEGFR family
|
- | 6-11 | - | - | - |
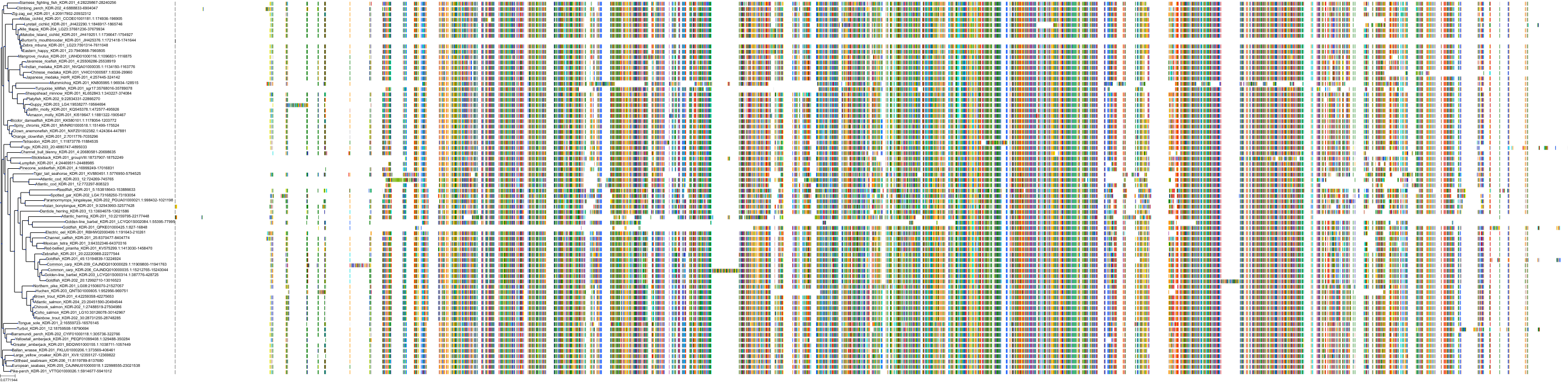
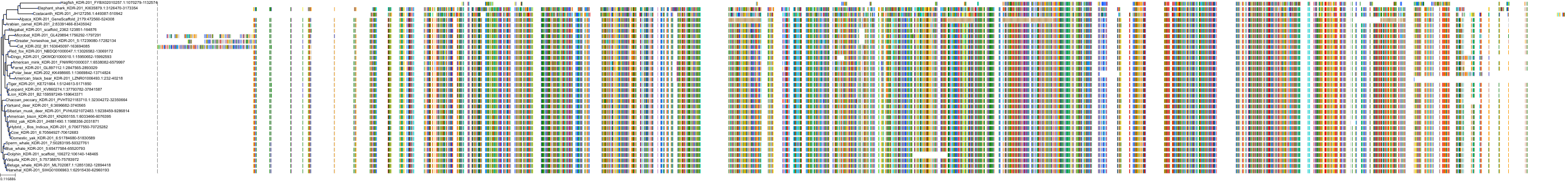
Target Conservation
|
Protein: Vascular endothelial growth factor receptor 2 Description: Vascular endothelial growth factor receptor 2 Organism : Homo sapiens P35968 ENSG00000128052 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEMBL | CHEMBL253969 |
| DrugBank | DB12962 |
| FDA SRS | W1B375O5M2 |
| Guide to Pharmacology | 7881 |
| PDB | BFF |
| SureChEMBL | SCHEMBL187261 |
| ZINC | ZINC000003834191 |
 Homo sapiens
Homo sapiens