Structure
| InChI Key | ZTFBIUXIQYRUNT-MDWZMJQESA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C25H23F3N4O2 |
| Molecular Weight | 468.48 |
| AlogP | 6.06 |
| Hydrogen Bond Acceptor | 6.0 |
| Number of Rotational Bond | 10.0 |
| Polar Surface Area | 65.97 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 4.0 |
| Heavy Atoms | 34.0 |
Pharmacology
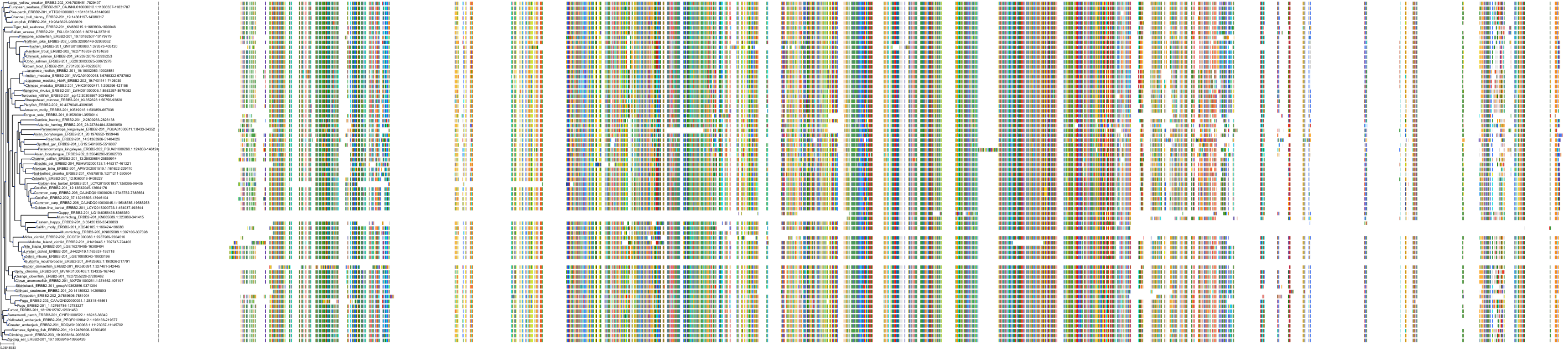
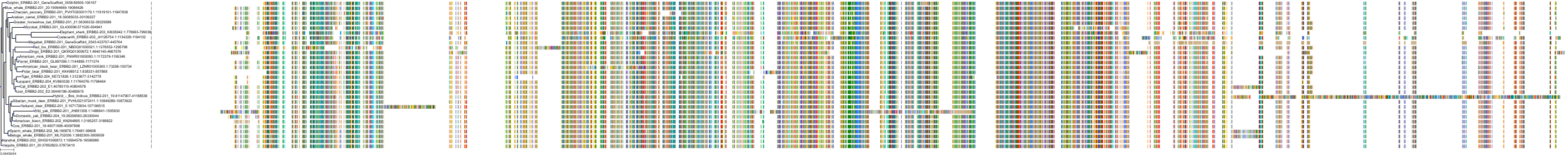
Target Conservation
|
Protein: Receptor protein-tyrosine kinase erbB-2 Description: Receptor tyrosine-protein kinase erbB-2 Organism : Homo sapiens P04626 ENSG00000141736 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEMBL | CHEMBL1614707 |
| DrugBank | DB12682 |
| FDA SRS | V734AZP9BR |
| Guide to Pharmacology | 6011 |
| PubChem | 6444692 |
| SureChEMBL | SCHEMBL94943 |
| ZINC | ZINC000011679877 |
 Bos taurus
Bos taurus
 Homo sapiens
Homo sapiens