| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| UNII | 1C6V77QF41 |
| EPA CompTox | DTXSID6026294 |
Structure
| InChI Key | QYSXJUFSXHHAJI-YRZJJWOYSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C27H44O |
| Molecular Weight | 384.65 |
| AlogP | 7.62 |
| Hydrogen Bond Acceptor | 1.0 |
| Hydrogen Bond Donor | 1.0 |
| Number of Rotational Bond | 6.0 |
| Polar Surface Area | 20.23 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 0.0 |
| Heavy Atoms | 28.0 |
Pharmacology
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Ion channel
Ligand-gated ion channel
Glycine receptor
|
400 | - | - | - | - | |
|
Transcription factor
Nuclear receptor
Nuclear hormone receptor subfamily 1
Nuclear hormone receptor subfamily 1 group I
Nuclear hormone receptor subfamily 1 group I member 1
|
0.21-900 | - | - | - | - | |
|
Unclassified protein
|
- | - | - | - | 74-80 |
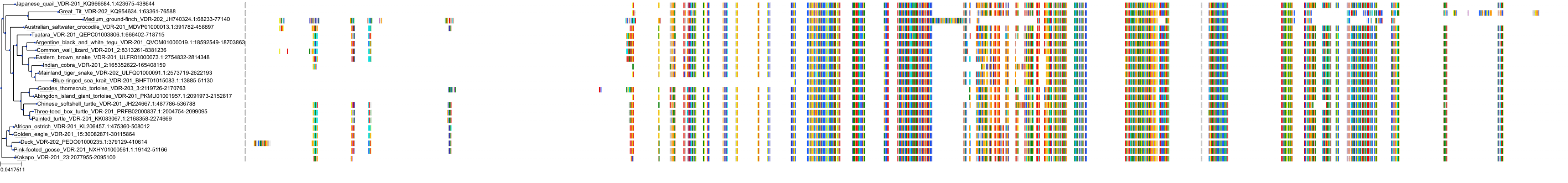
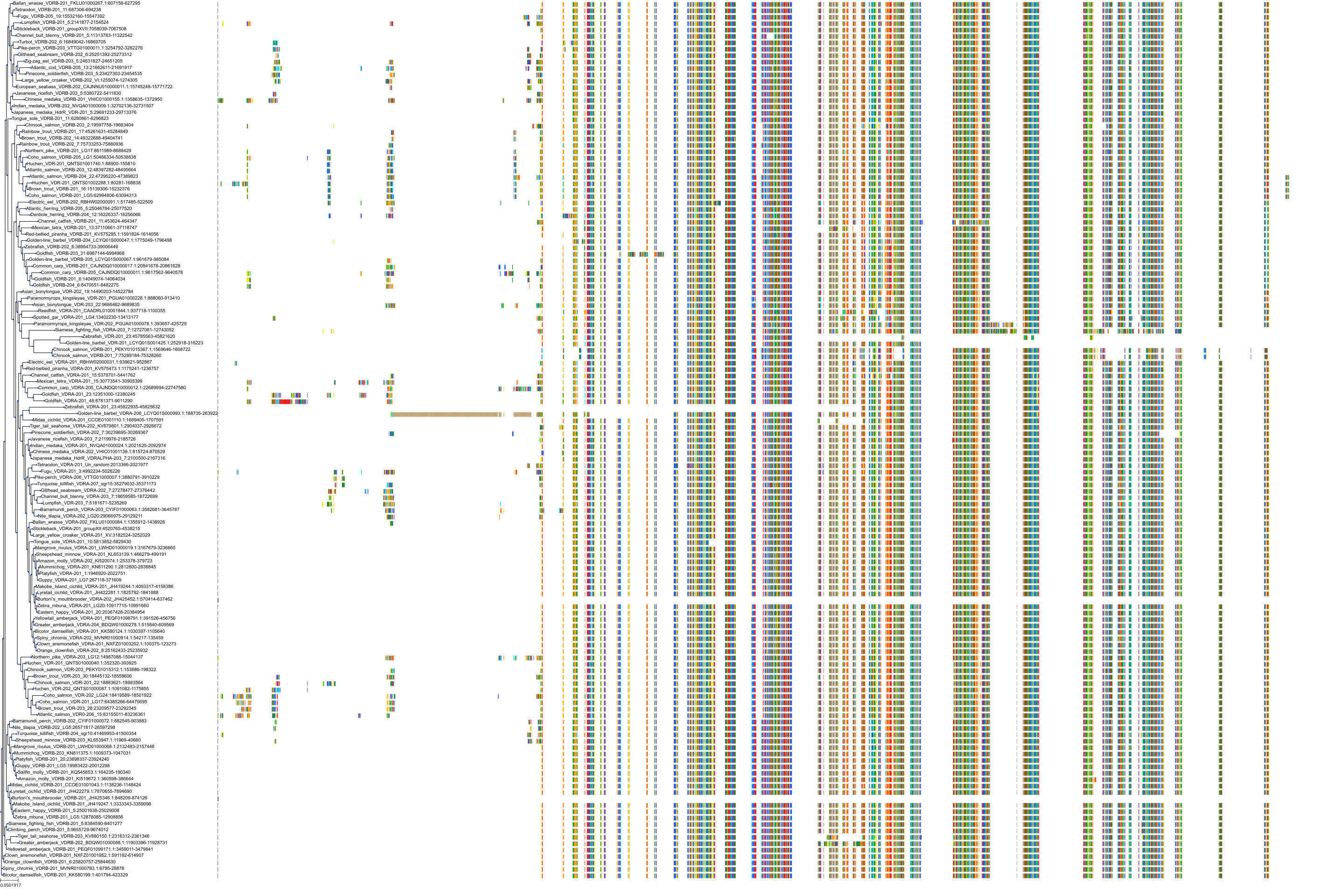
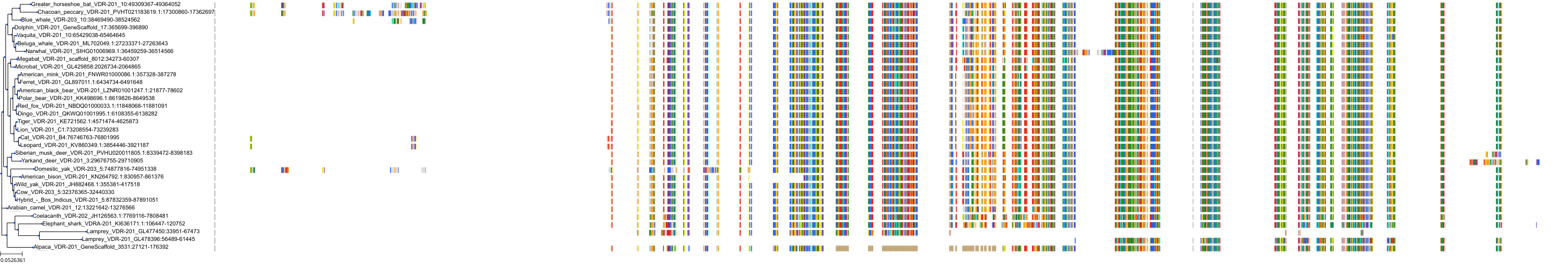
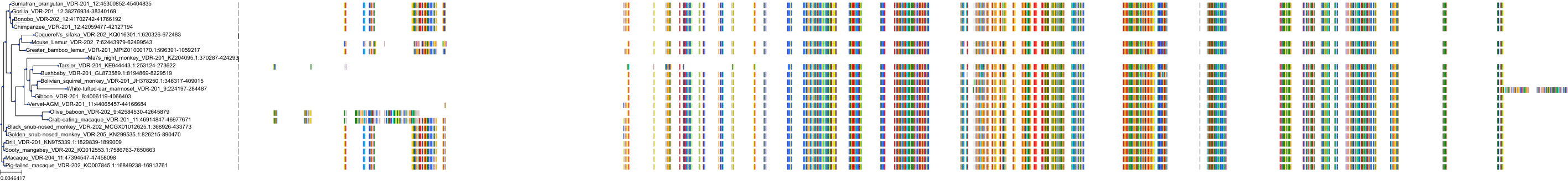
Target Conservation
|
Protein: Vitamin D receptor Description: Vitamin D3 receptor Organism : Homo sapiens P11473 ENSG00000111424 |
||||
Related Entries
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 28940 |
| ChEMBL | CHEMBL1042 |
| DrugBank | DB00169 |
| DrugCentral | 2840 |
| FDA SRS | 1C6V77QF41 |
| Human Metabolome Database | HMDB0000876 |
| KEGG | C05443 |
| PubChem | 5283710 |
| SureChEMBL | SCHEMBL3126 |
| ZINC | ZINC000004474460 |
 Escherichia coli
Escherichia coli
 Homo sapiens
Homo sapiens
 Mus musculus
Mus musculus
 Rattus norvegicus
Rattus norvegicus