Structure
| InChI Key | JGRXMPYUTJLTKT-UHFFFAOYSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C14H11ClN2O4 |
| Molecular Weight | 306.71 |
| AlogP | 1.92 |
| Hydrogen Bond Acceptor | 4.0 |
| Hydrogen Bond Donor | 3.0 |
| Number of Rotational Bond | 4.0 |
| Polar Surface Area | 99.52 |
| Molecular species | ACID |
| Aromatic Rings | 2.0 |
| Heavy Atoms | 21.0 |
Pharmacology
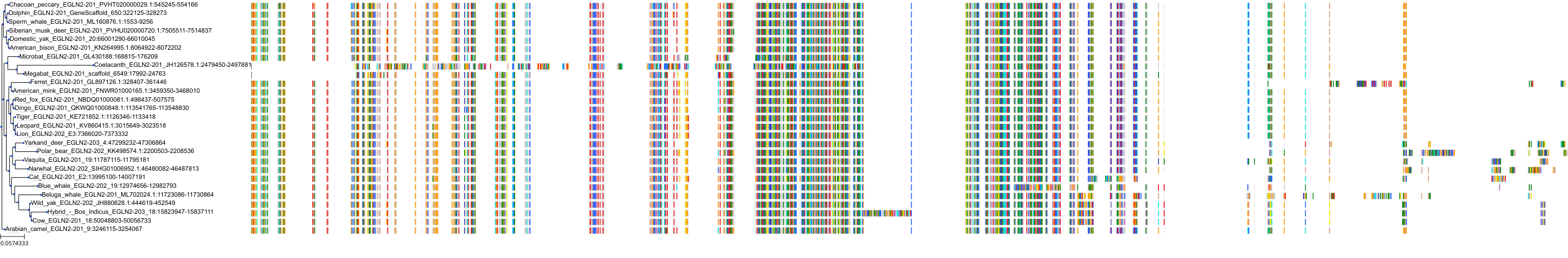
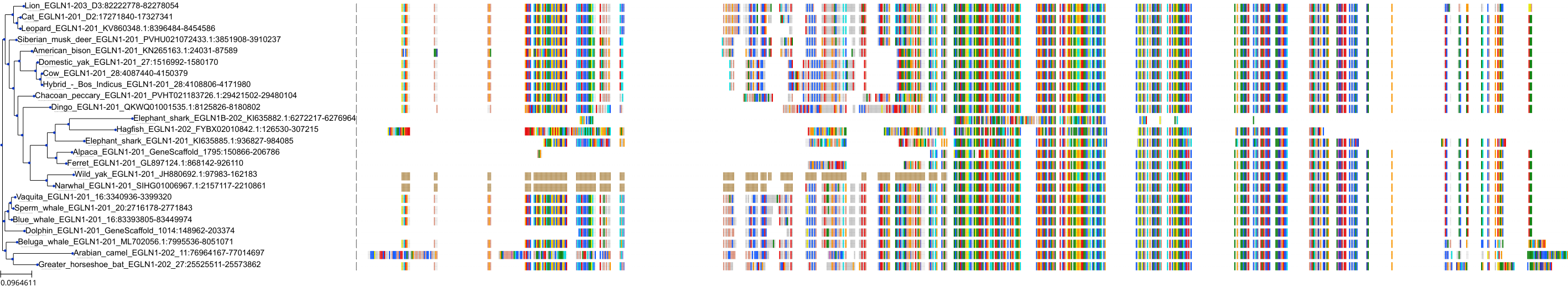
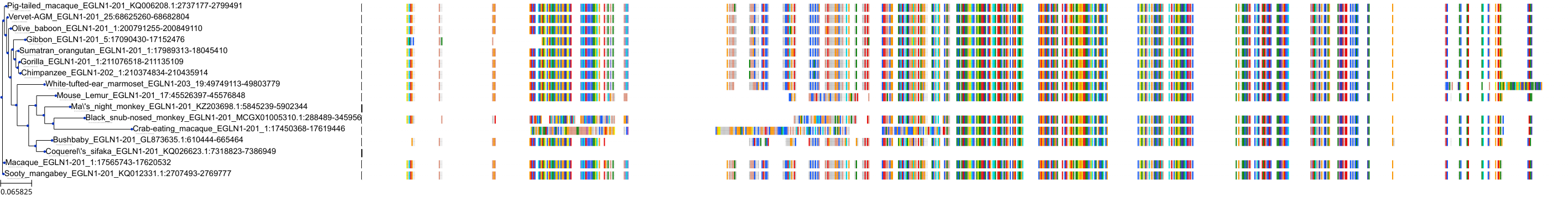
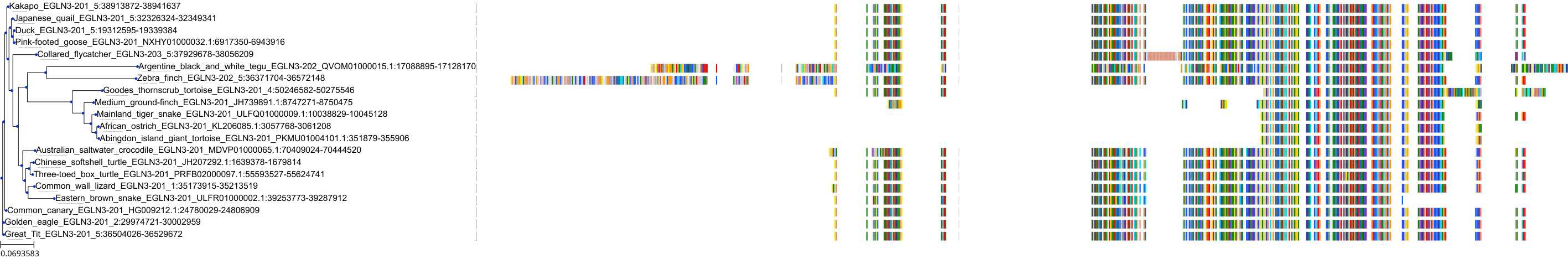
Target Conservation
|
Protein: Hypoxia-inducible factor prolyl hydroxylase 1 Description: Prolyl hydroxylase EGLN2 Organism : Homo sapiens Q96KS0 ENSG00000269858 |
||||
|
Protein: Egl nine homolog 1 Description: Egl nine homolog 1 Organism : Homo sapiens Q9GZT9 ENSG00000135766 |
||||
|
Protein: Egl nine homolog 3 Description: Prolyl hydroxylase EGLN3 Organism : Homo sapiens Q9H6Z9 ENSG00000129521 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEMBL | CHEMBL3646221 |
| DrugBank | DB12255 |
| FDA SRS | I60W9520VV |
| PDB | A1Z |
| PubChem | 23634441 |
| SureChEMBL | SCHEMBL1920738 |
| ZINC | ZINC000117532869 |
 Homo sapiens
Homo sapiens
 Paramecium bursaria Chlorella virus 1
Paramecium bursaria Chlorella virus 1