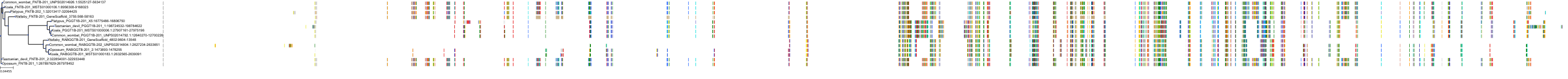| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| UNII | MAT637500A |
| EPA CompTox | DTXSID5041140 |
Structure
| InChI Key | PLHJCIYEEKOWNM-HHHXNRCGSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C27H22Cl2N4O |
| Molecular Weight | 489.41 |
| AlogP | 5.5 |
| Hydrogen Bond Acceptor | 5.0 |
| Hydrogen Bond Donor | 1.0 |
| Number of Rotational Bond | 4.0 |
| Polar Surface Area | 65.84 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 5.0 |
| Heavy Atoms | 34.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Protein farnesyltransferase inhibitor | INHIBITOR | PubMed |
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Enzyme
Transferase
|
1.6-100 | 0.57-7.9 | - | - | 1.6 | |
|
Enzyme
|
1.6-100 | 0.57-7.9 | - | - | 1.6 |
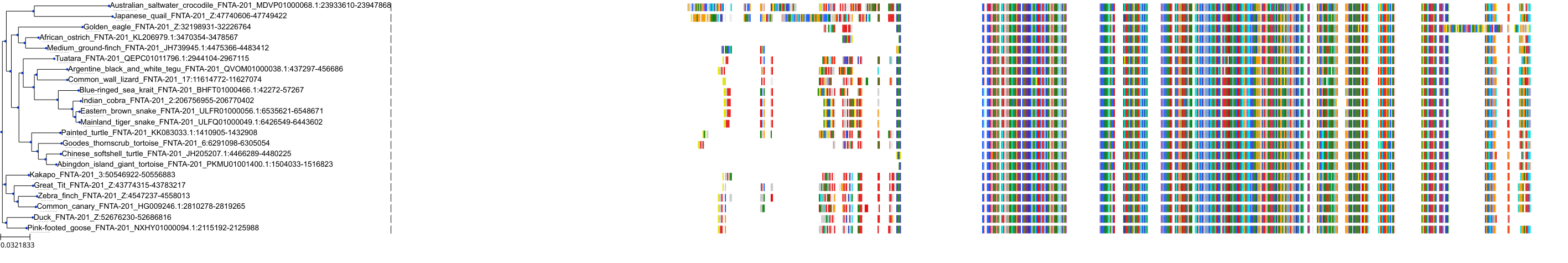
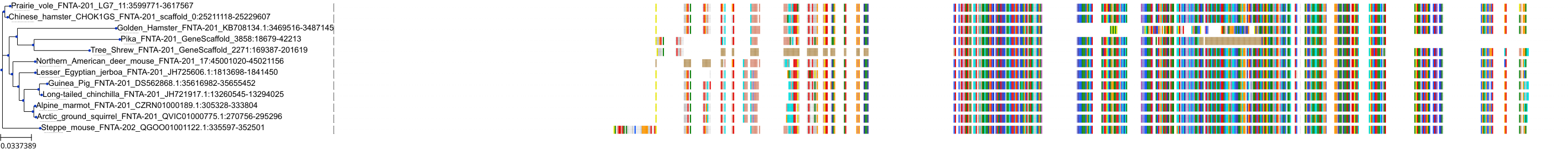
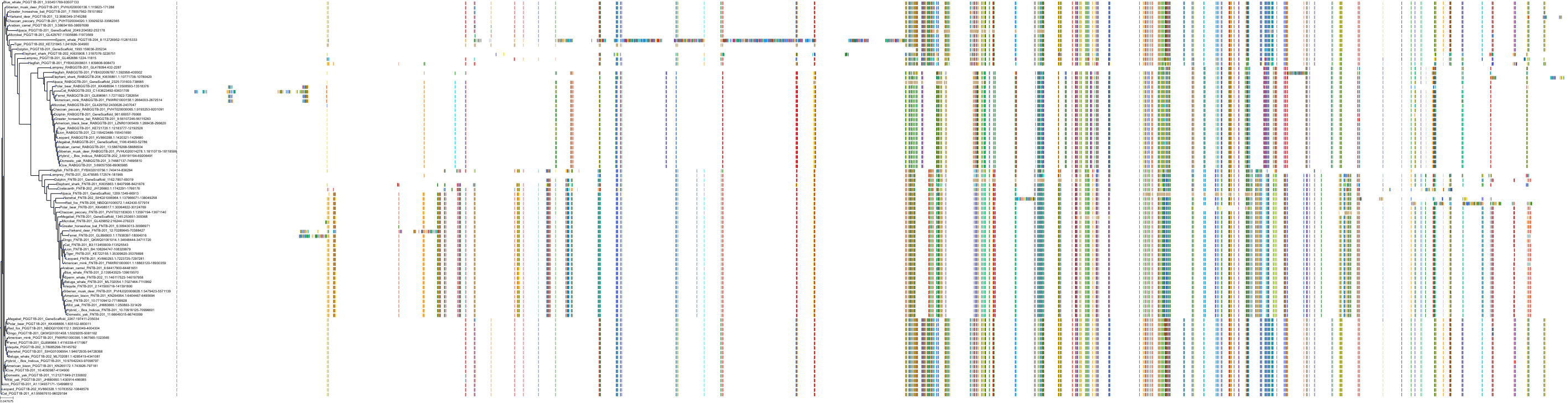
Target Conservation
|
Protein: Protein farnesyltransferase Description: Protein farnesyltransferase/geranylgeranyltransferase type-1 subunit alpha Organism : Homo sapiens P49354 ENSG00000168522 |
||||
|
Protein: Protein farnesyltransferase Description: Protein farnesyltransferase subunit beta Organism : Homo sapiens P49356 ENSG00000257365 |
||||
Related Entries
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 141969 |
| ChEMBL | CHEMBL289228 |
| DrugBank | DB04960 |
| FDA SRS | MAT637500A |
| Guide to Pharmacology | 8025 |
| PDB | JAN |
| PubChem | 159324 |
| SureChEMBL | SCHEMBL21544535 |
| ZINC | ZINC000024809155 |
 Bos taurus
Bos taurus
 Homo sapiens
Homo sapiens
 Mus musculus
Mus musculus
 Plasmodium falciparum
Plasmodium falciparum
 Rattus norvegicus
Rattus norvegicus
 Trypanosoma cruzi
Trypanosoma cruzi