Structure
| InChI Key | OAVGBZOFDPFGPJ-UHFFFAOYSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C25H22N6O2 |
| Molecular Weight | 438.49 |
| AlogP | 2.43 |
| Hydrogen Bond Acceptor | 6.0 |
| Hydrogen Bond Donor | 2.0 |
| Number of Rotational Bond | 3.0 |
| Polar Surface Area | 94.22 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 4.0 |
| Heavy Atoms | 33.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Protein kinase C (PKC) inhibitor | INHIBITOR | PubMed |
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Enzyme
Kinase
Protein Kinase
AGC protein kinase group
AGC protein kinase PKC family
AGC protein kinase PKC alpha subfamily
|
- | 2-234 | - | - | - | |
|
Enzyme
Kinase
Protein Kinase
AGC protein kinase group
AGC protein kinase PKC family
AGC protein kinase PKC delta subfamily
|
- | 1-81 | - | - | - | |
|
Enzyme
Kinase
Protein Kinase
AGC protein kinase group
AGC protein kinase PKC family
AGC protein kinase PKC eta subfamily
|
- | 6.1-6.2 | - | - | - | |
|
Enzyme
Kinase
Protein Kinase
CAMK protein kinase group
CAMK protein kinase PIM family
|
- | 50.12 | - | - | - | |
|
Enzyme
Kinase
Protein Kinase
CMGC protein kinase group
CMGC protein kinase GSK family
|
- | 870 | - | - | - |
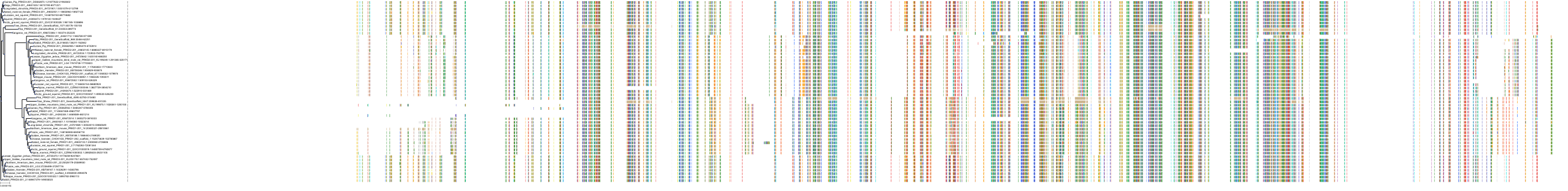
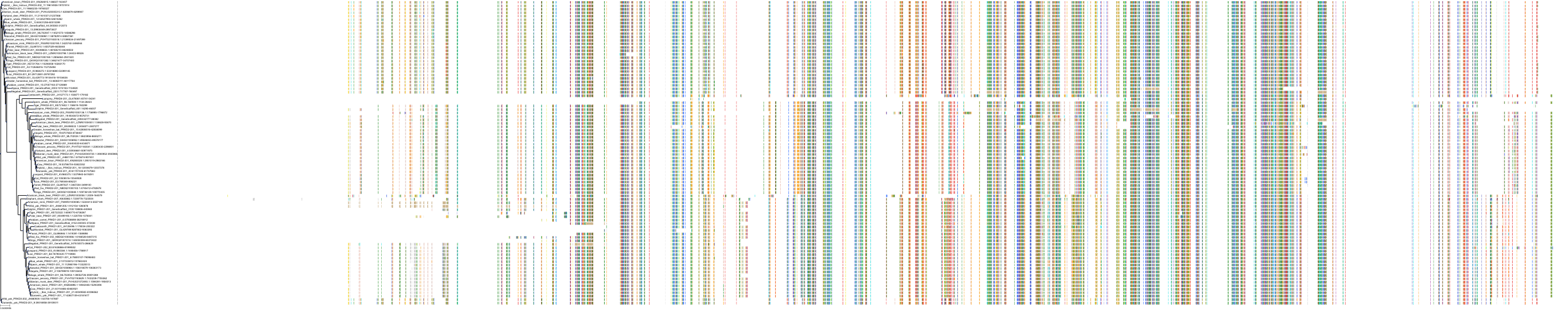
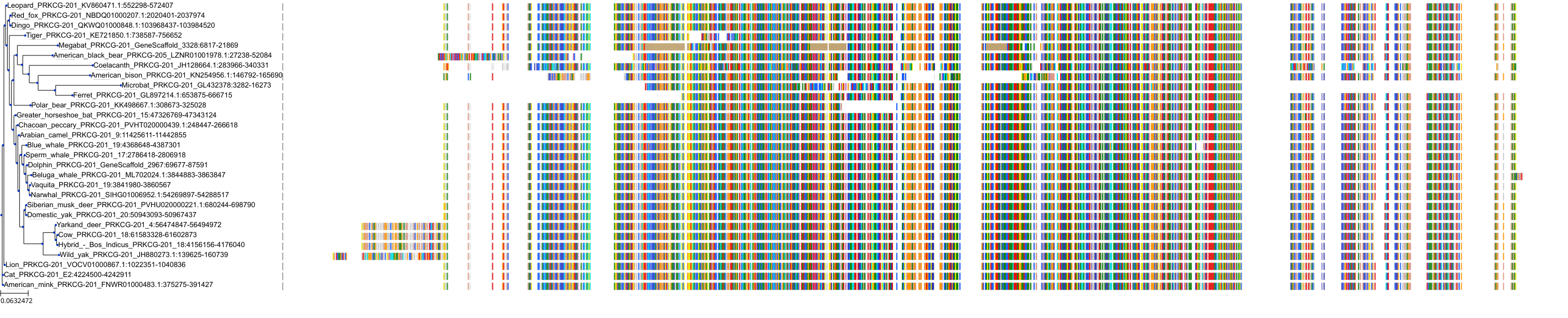
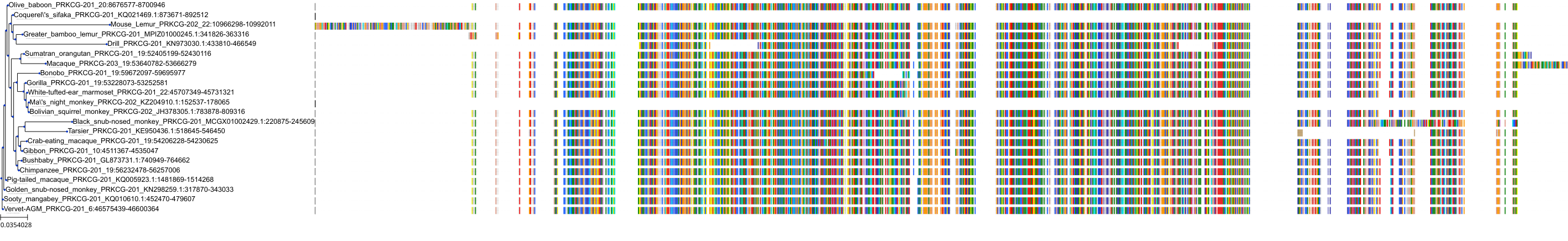
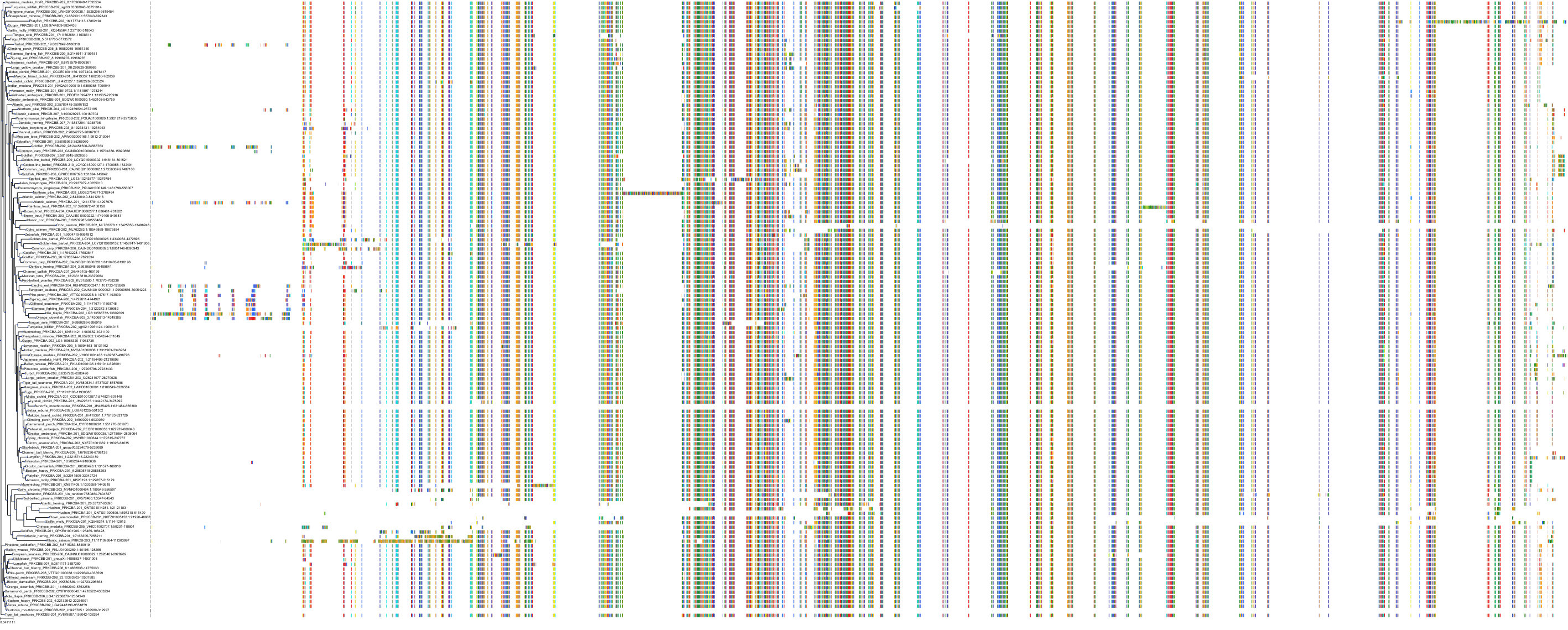
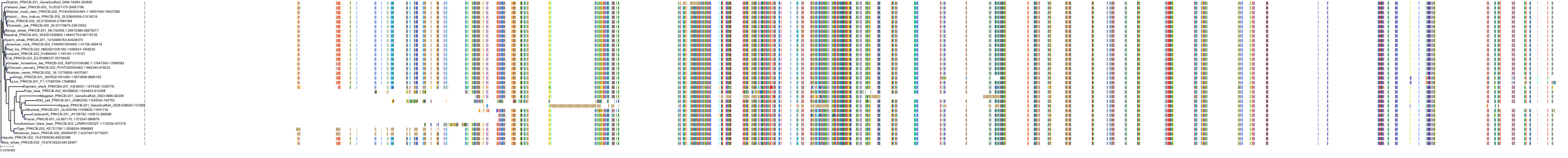
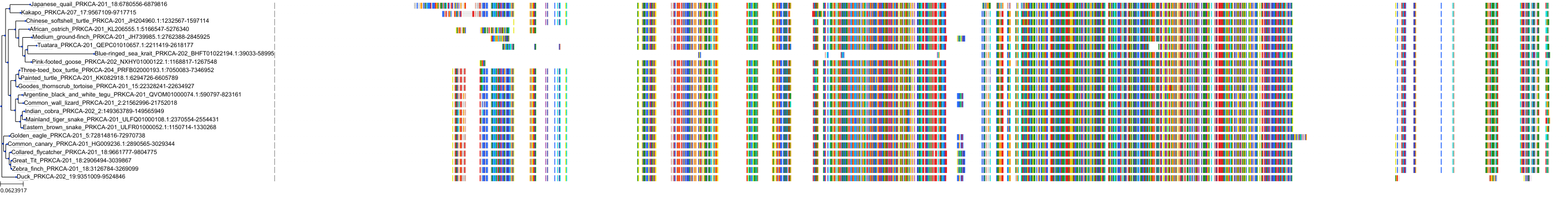
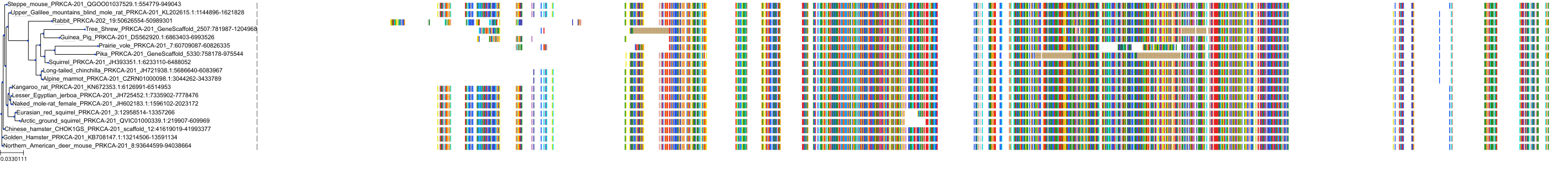
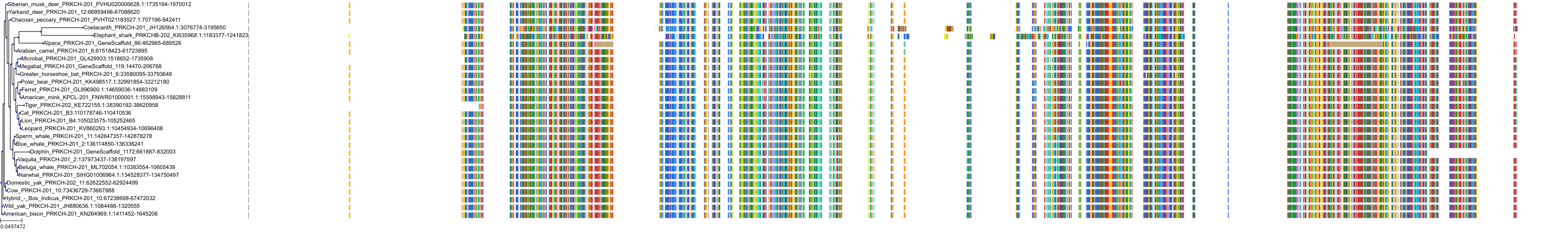
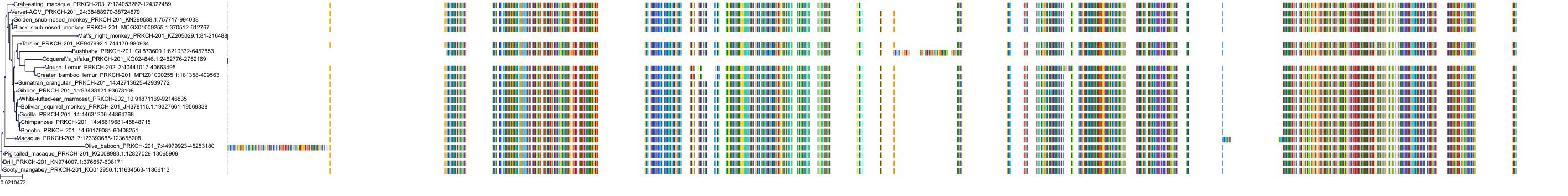
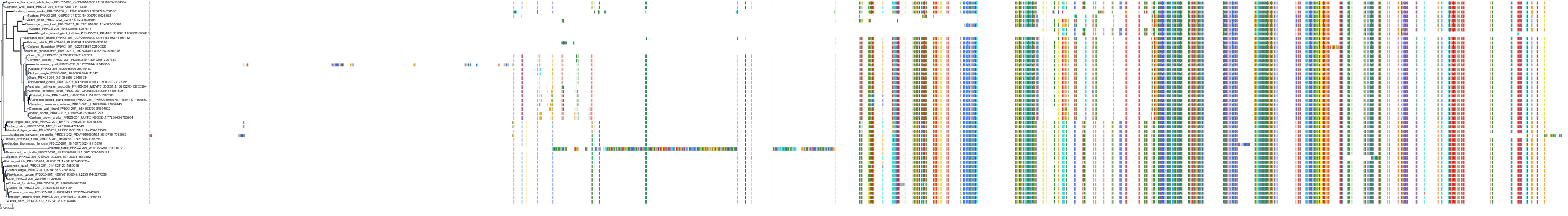
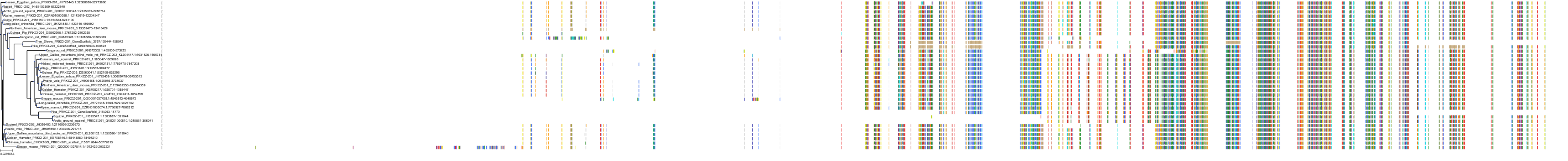
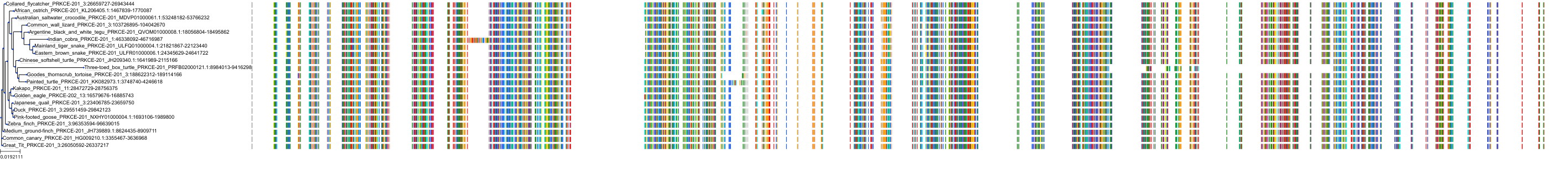
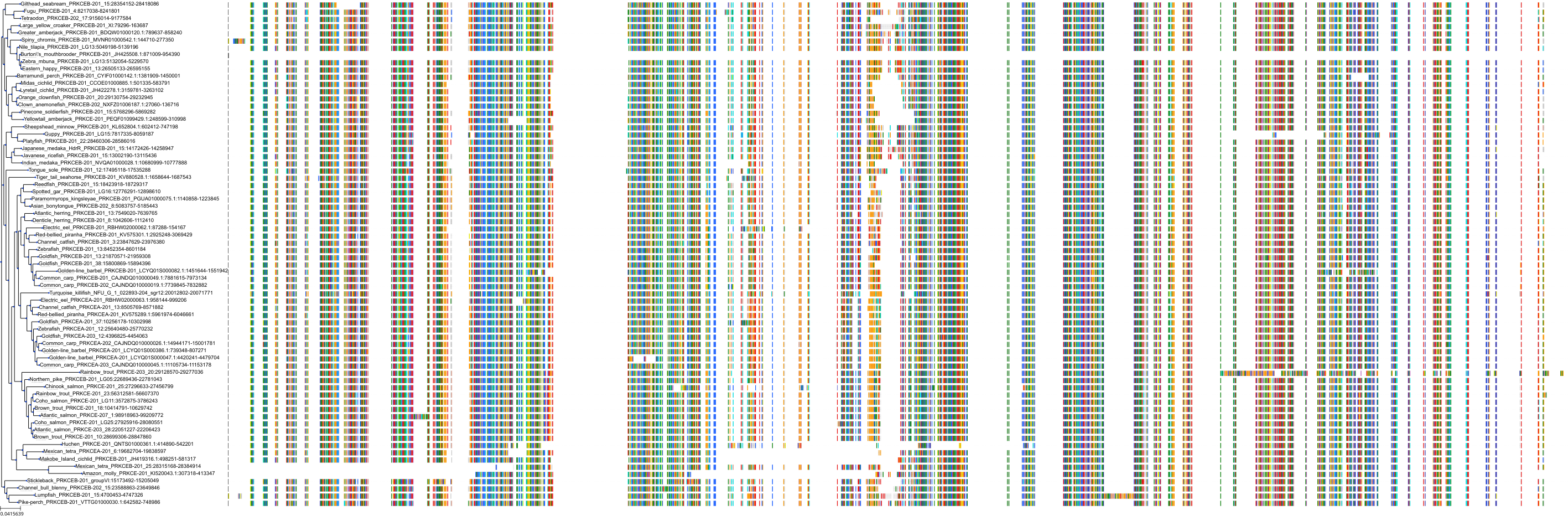
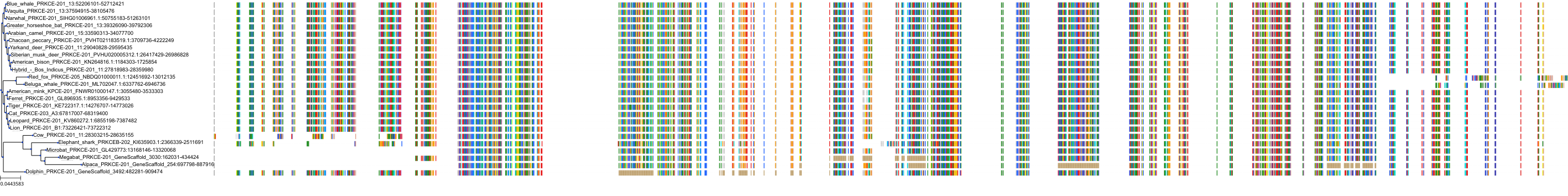
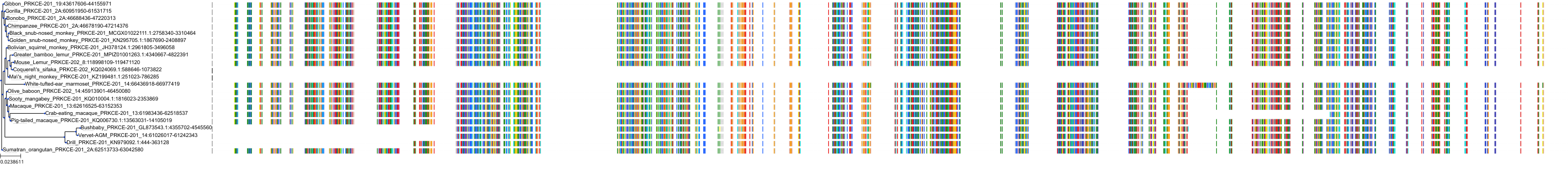
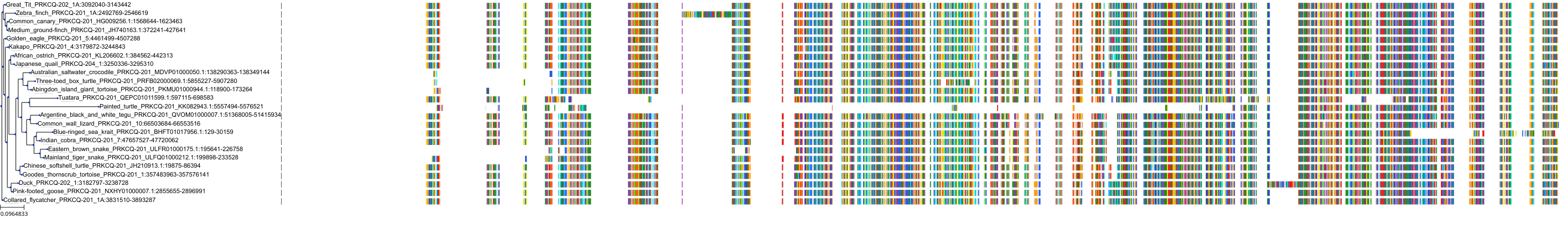
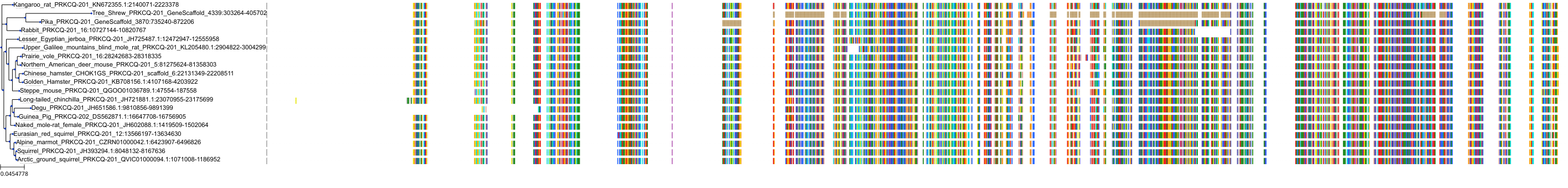
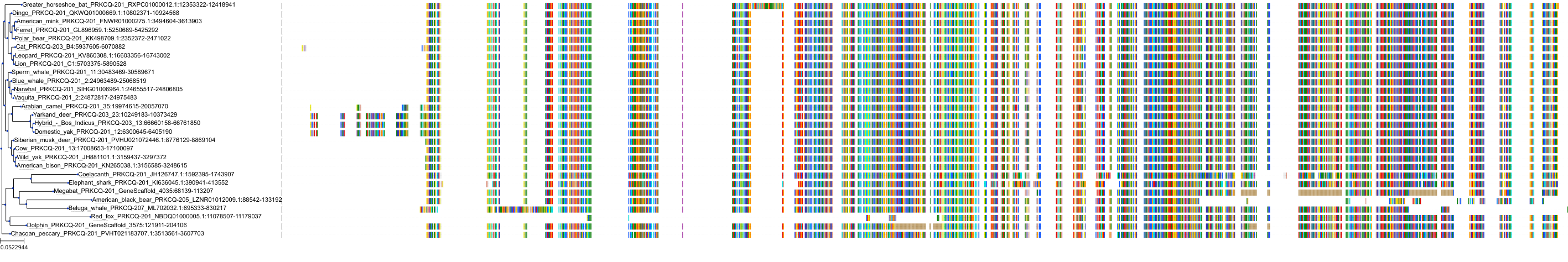
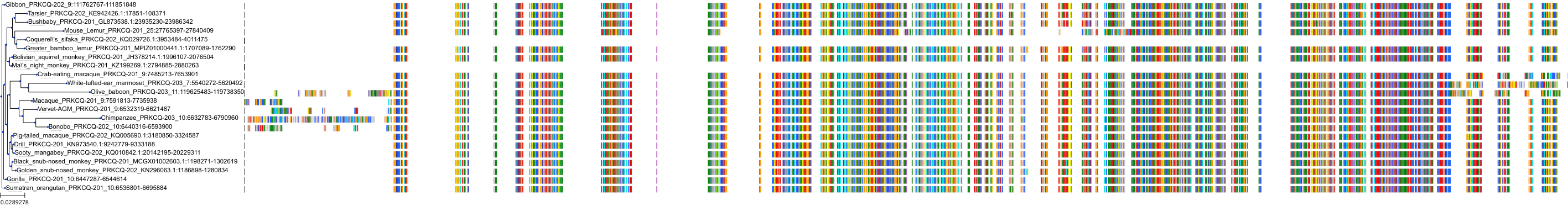
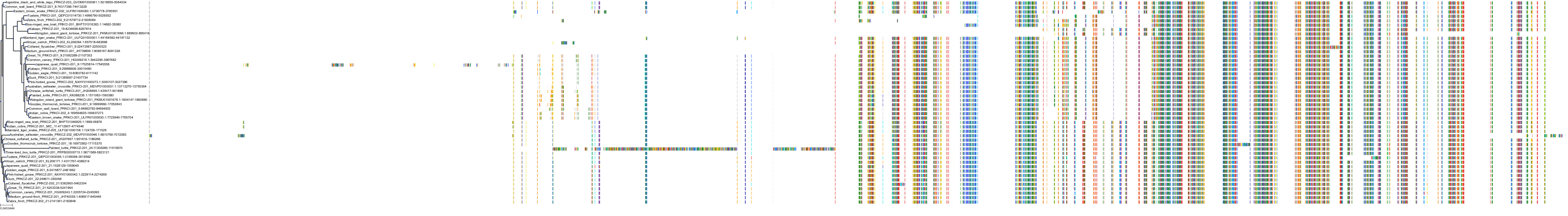
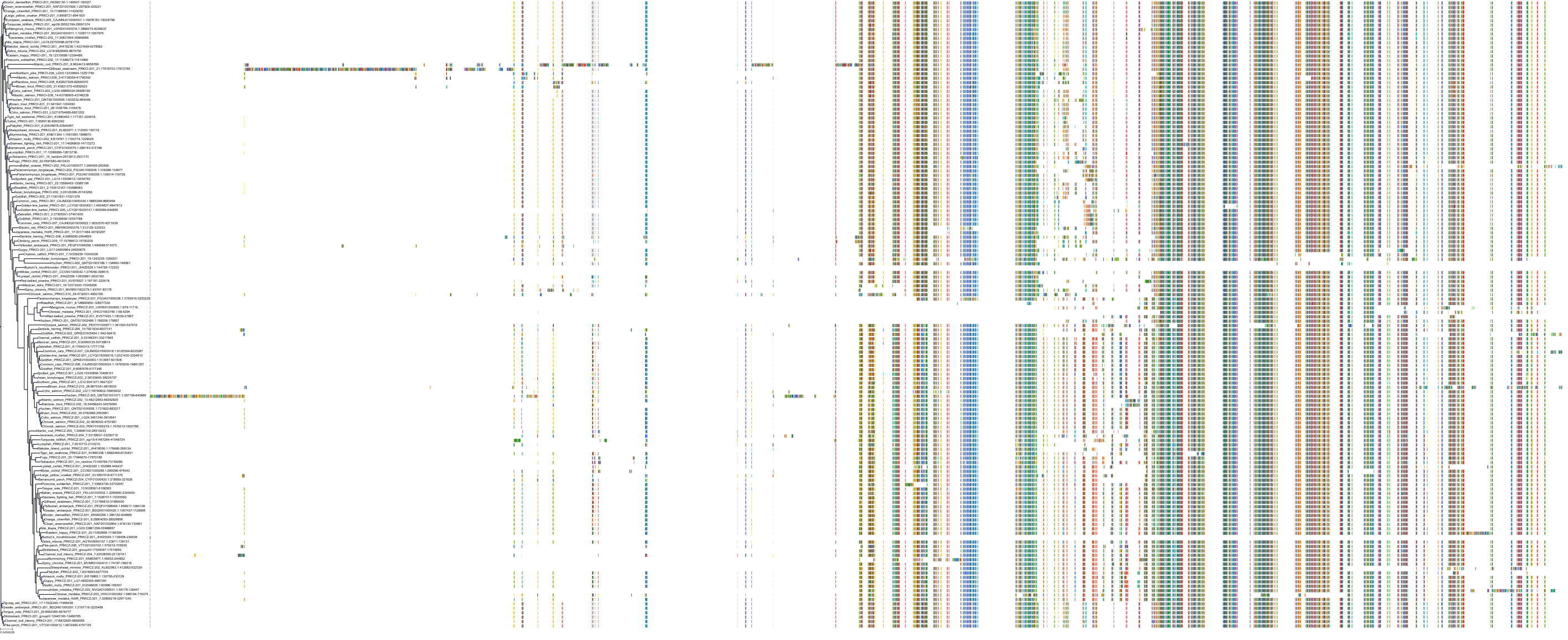
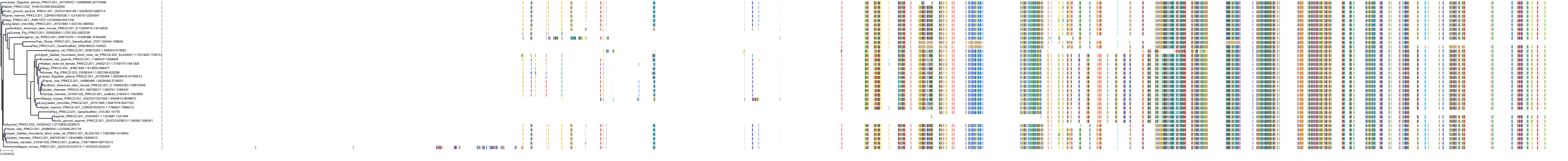
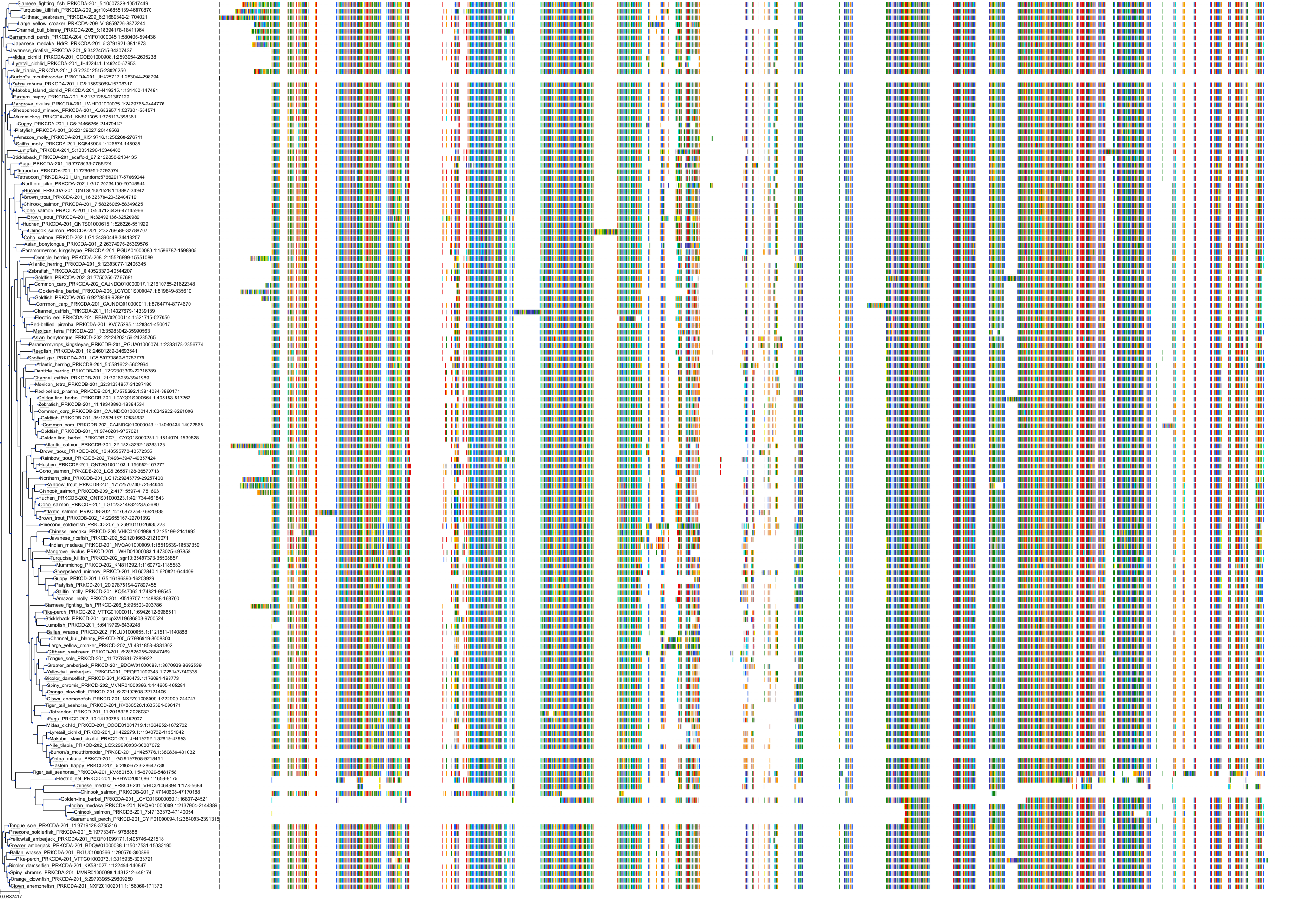
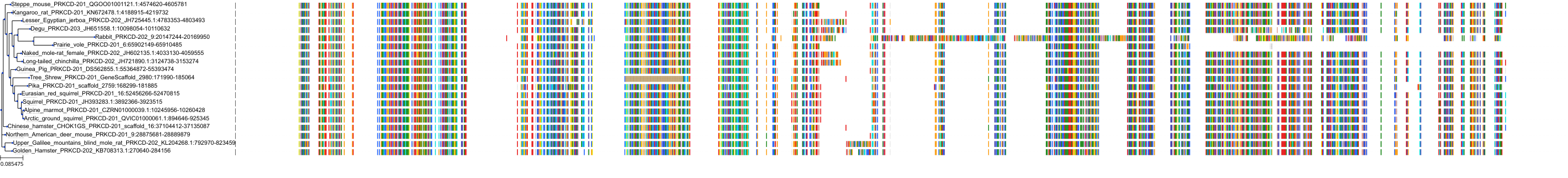
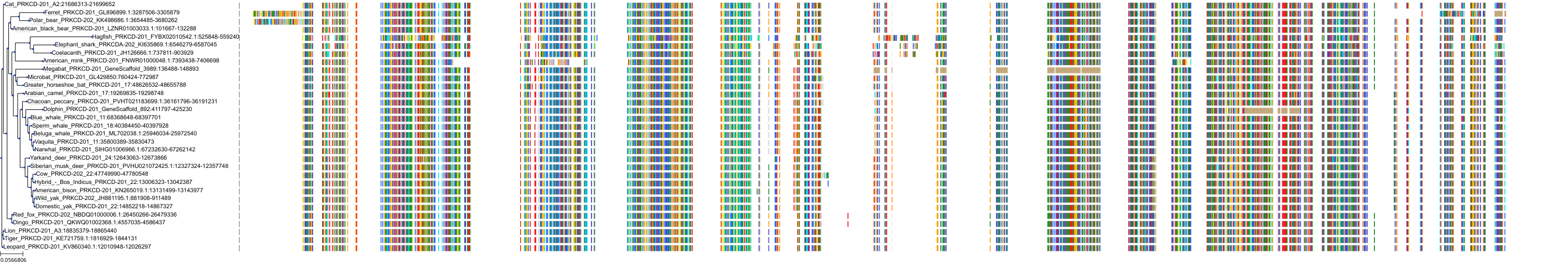
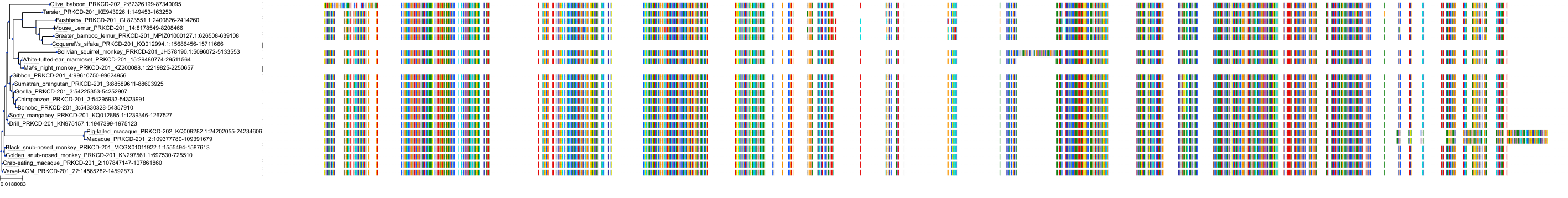
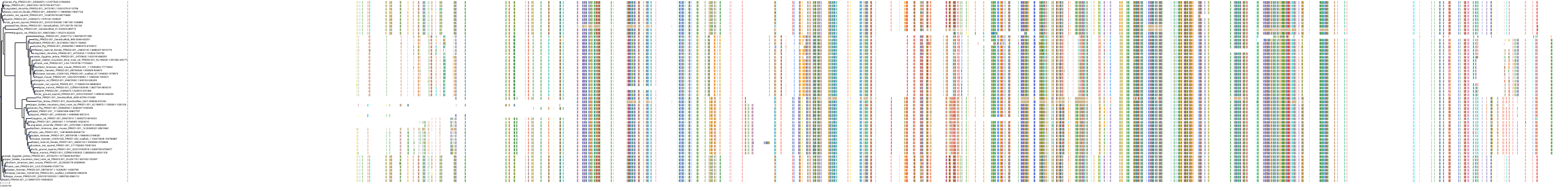
Target Conservation
|
Protein: Protein kinase C (PKC) Description: Serine/threonine-protein kinase D3 Organism : Homo sapiens O94806 ENSG00000115825 |
||||
|
Protein: Protein kinase C (PKC) Description: Protein kinase C gamma type Organism : Homo sapiens P05129 ENSG00000126583 |
||||
|
Protein: Protein kinase C (PKC) Description: Protein kinase C beta type Organism : Homo sapiens P05771 ENSG00000166501 |
||||
|
Protein: Protein kinase C (PKC) Description: Protein kinase C alpha type Organism : Homo sapiens P17252 ENSG00000154229 |
||||
|
Protein: Protein kinase C (PKC) Description: Protein kinase C eta type Organism : Homo sapiens P24723 ENSG00000027075 |
||||
|
Protein: Protein kinase C (PKC) Description: Protein kinase C iota type Organism : Homo sapiens P41743 ENSG00000163558 |
||||
|
Protein: Protein kinase C (PKC) Description: Protein kinase C epsilon type Organism : Homo sapiens Q02156 ENSG00000171132 |
||||
|
Protein: Protein kinase C (PKC) Description: Protein kinase C theta type Organism : Homo sapiens Q04759 ENSG00000065675 |
||||
|
Protein: Protein kinase C (PKC) Description: Protein kinase C zeta type Organism : Homo sapiens Q05513 ENSG00000067606 |
||||
|
Protein: Protein kinase C (PKC) Description: Protein kinase C delta type Organism : Homo sapiens Q05655 ENSG00000163932 |
||||
|
Protein: Protein kinase C (PKC) Description: Serine/threonine-protein kinase D1 Organism : Homo sapiens Q15139 ENSG00000184304 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 90531 |
| ChEMBL | CHEMBL565612 |
| DrugBank | DB12369 |
| FDA SRS | 7I279E1NZ8 |
| Guide to Pharmacology | 7946 |
| PDB | LW4 |
| PubChem | 10296883 |
| SureChEMBL | SCHEMBL2500835 |
| ZINC | ZINC000003973984 |
 Homo sapiens
Homo sapiens
 Mus musculus
Mus musculus