| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| ATC | L01EE04 |
| UNII | 6UH91I579U |
| EPA CompTox | DTXSID3048944 |
Structure
| InChI Key | CYOHGALHFOKKQC-UHFFFAOYSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C17H15BrClFN4O3 |
| Molecular Weight | 457.69 |
| AlogP | 3.53 |
| Hydrogen Bond Acceptor | 6.0 |
| Hydrogen Bond Donor | 3.0 |
| Number of Rotational Bond | 6.0 |
| Polar Surface Area | 88.41 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 3.0 |
| Heavy Atoms | 27.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Dual specificity mitogen-activated protein kinase kinase 1 inhibitor | INHIBITOR | PubMed PubMed |
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Enzyme
Kinase
Protein Kinase
STE protein kinase group
STE protein kinase STE7 family
|
- | 9-80 | 99-530 | - | - |
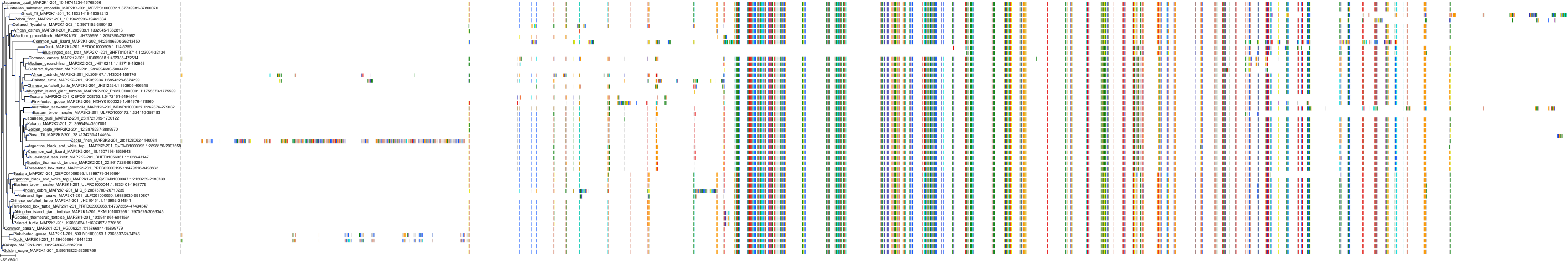
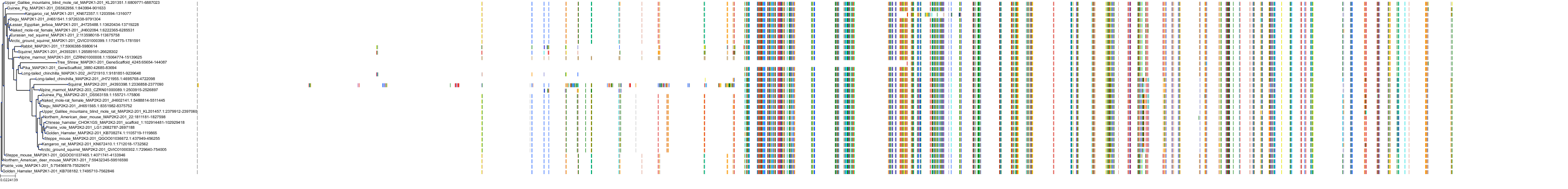
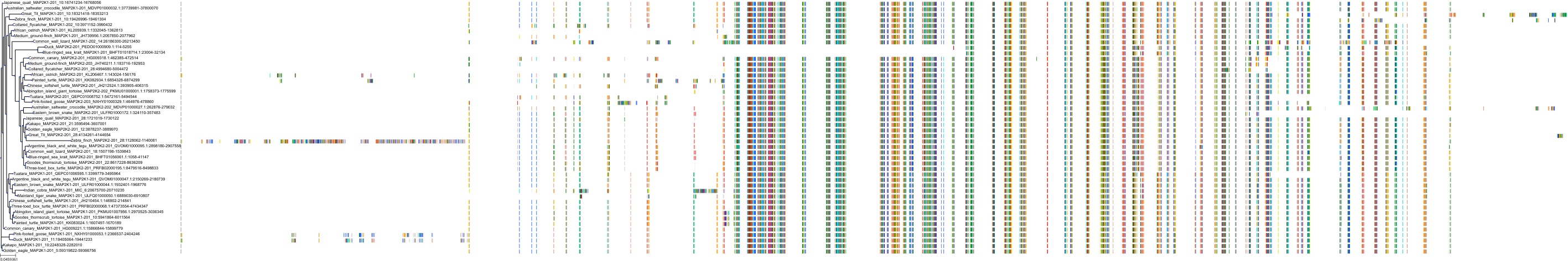
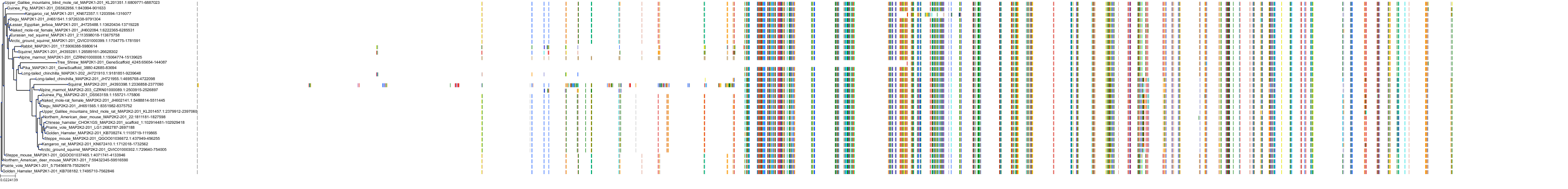
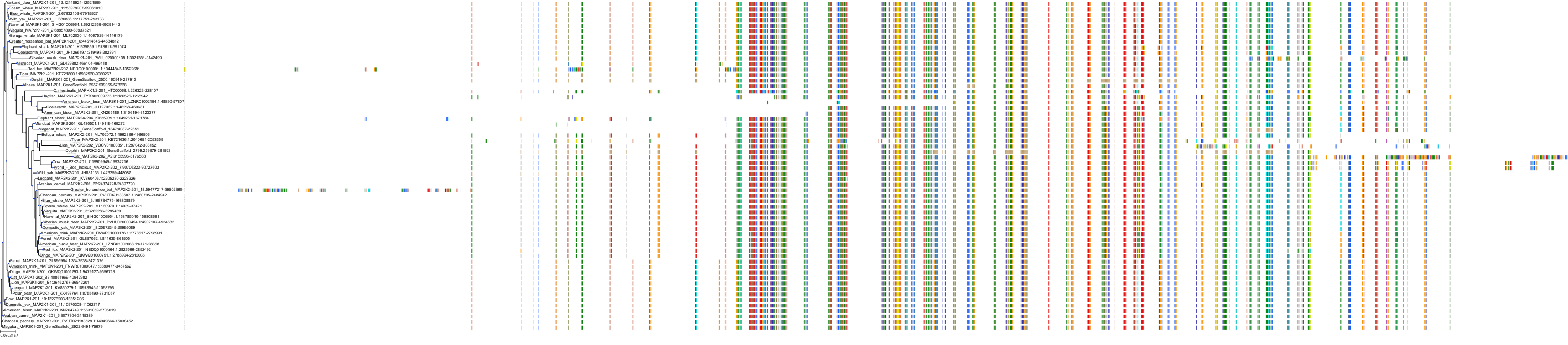
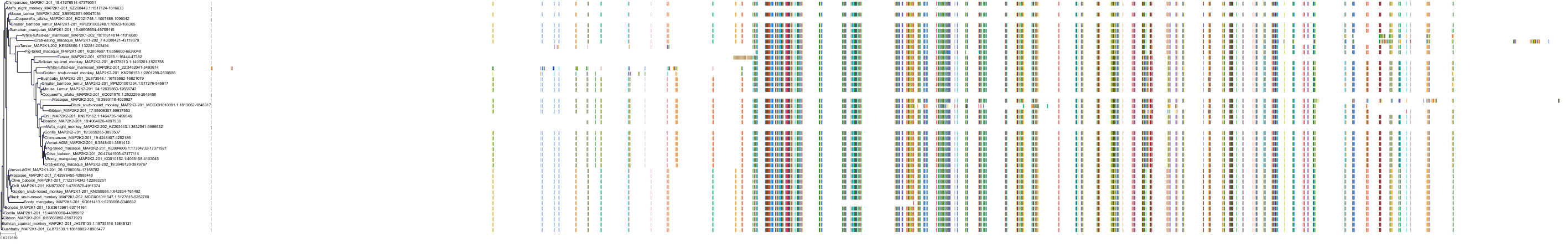
Target Conservation
|
Protein: Dual specificity mitogen-activated protein kinase kinase 2 Description: Dual specificity mitogen-activated protein kinase kinase 2 Organism : Homo sapiens P36507 ENSG00000126934 |
||||
|
Protein: Dual specificity mitogen-activated protein kinase kinase 1 Description: Dual specificity mitogen-activated protein kinase kinase 1 Organism : Homo sapiens Q02750 ENSG00000169032 |
||||
Related Entries
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 90227 |
| ChEMBL | CHEMBL1614701 |
| DrugBank | DB11689 |
| DrugCentral | 5388 |
| FDA SRS | 6UH91I579U |
| Guide to Pharmacology | 5665 |
| PDB | 3EW |
| PharmGKB | PA166129529 |
| PubChem | 10127622 |
| SureChEMBL | SCHEMBL155456 |
| ZINC | ZINC000031773258 |
 Homo sapiens
Homo sapiens
 Mus musculus
Mus musculus