Structure
| InChI Key | PJBFVWGQFLYWCB-UHFFFAOYSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C20H28N4O2 |
| Molecular Weight | 356.47 |
| AlogP | 2.78 |
| Hydrogen Bond Acceptor | 5.0 |
| Hydrogen Bond Donor | 1.0 |
| Number of Rotational Bond | 5.0 |
| Polar Surface Area | 72.68 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 2.0 |
| Heavy Atoms | 26.0 |
Pharmacology
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Membrane receptor
Family A G protein-coupled receptor
Small molecule receptor (family A GPCR)
Nucleotide-like receptor (family A GPCR)
Adenosine receptor
|
- | - | - | 0.145-510 | - |
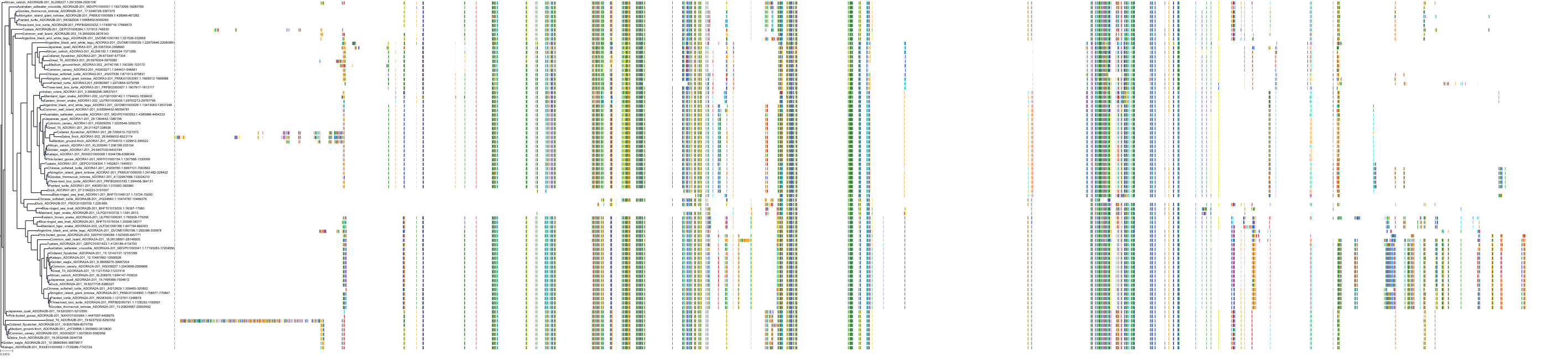
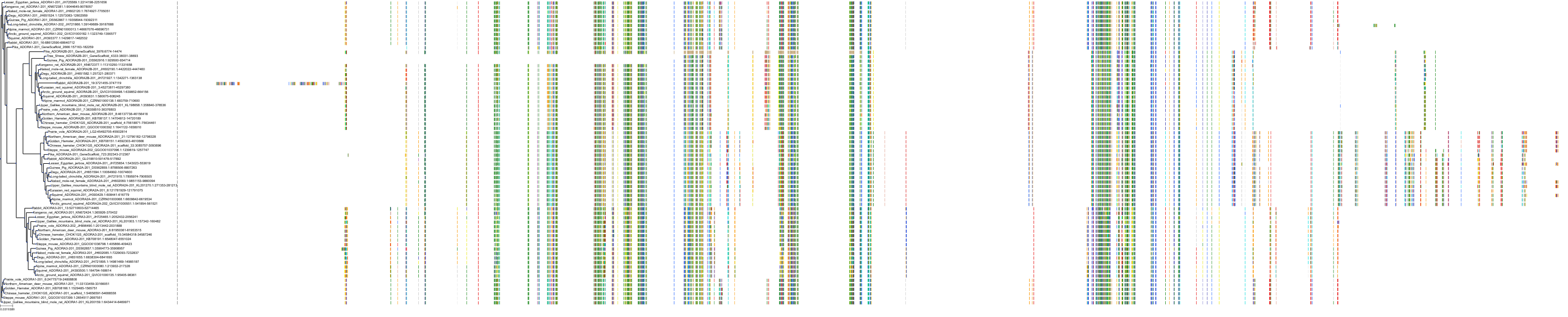
Target Conservation
|
Protein: Adenosine A1 receptor Description: Adenosine receptor A1 Organism : Homo sapiens P30542 ENSG00000163485 |
||||
Related Entries
Cross References
| Resources | Reference |
|---|---|
| ChEMBL | CHEMBL52333 |
| FDA SRS | 7805S5HIHX |
| Guide to Pharmacology | 5604 |
| PubChem | 11948288 |
| SureChEMBL | SCHEMBL18029490 |
 Cavia porcellus
Cavia porcellus
 Homo sapiens
Homo sapiens
 Mus musculus
Mus musculus
 Rattus norvegicus
Rattus norvegicus