| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| UNII | JPX07AFM0N |
| EPA CompTox | DTXSID40238961 |
Structure
| InChI Key | RDSACQWTXKSHJT-NSHDSACASA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C19H20F3IN2O5S |
| Molecular Weight | 572.34 |
| AlogP | 3.48 |
| Hydrogen Bond Acceptor | 6.0 |
| Hydrogen Bond Donor | 4.0 |
| Number of Rotational Bond | 9.0 |
| Polar Surface Area | 107.89 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 2.0 |
| Heavy Atoms | 31.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Dual specificity mitogen-activated protein kinase kinase 1 inhibitor | INHIBITOR | PubMed |
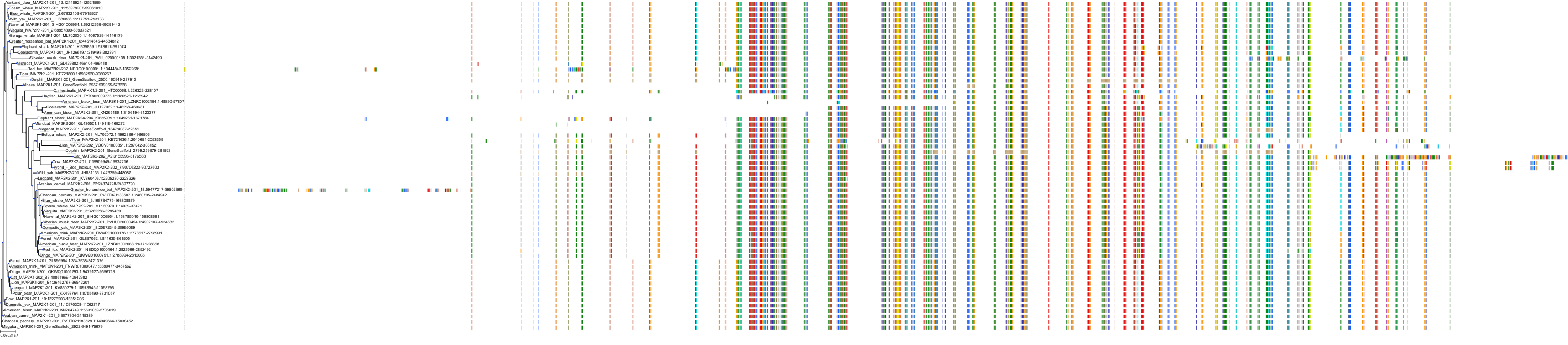
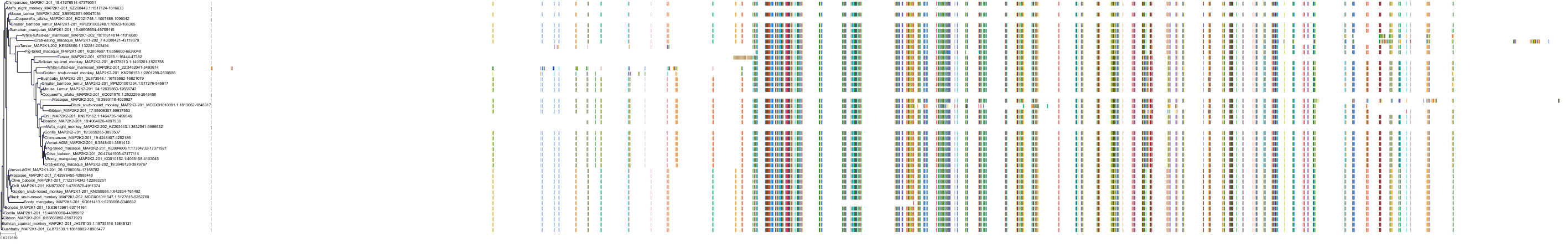
Target Conservation
|
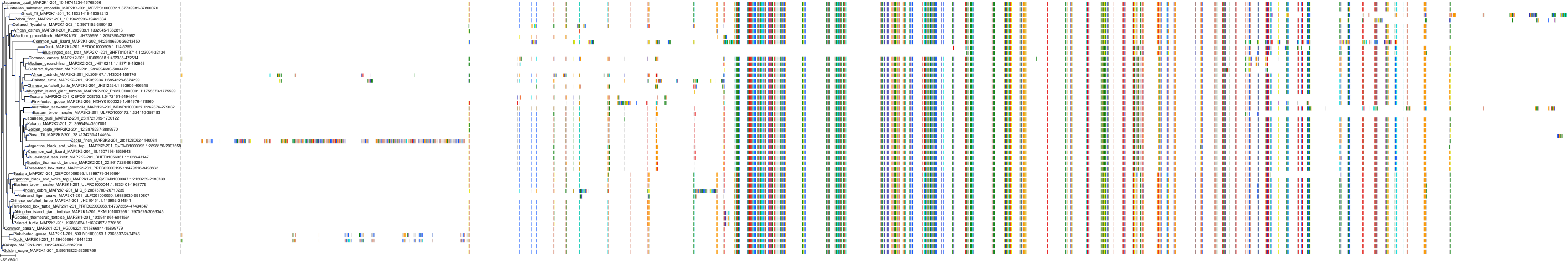
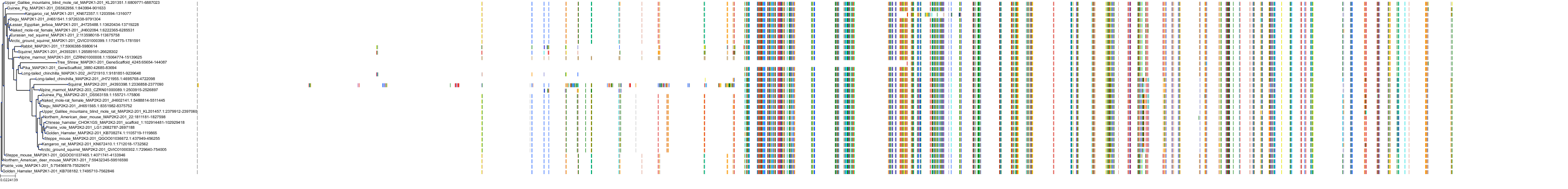
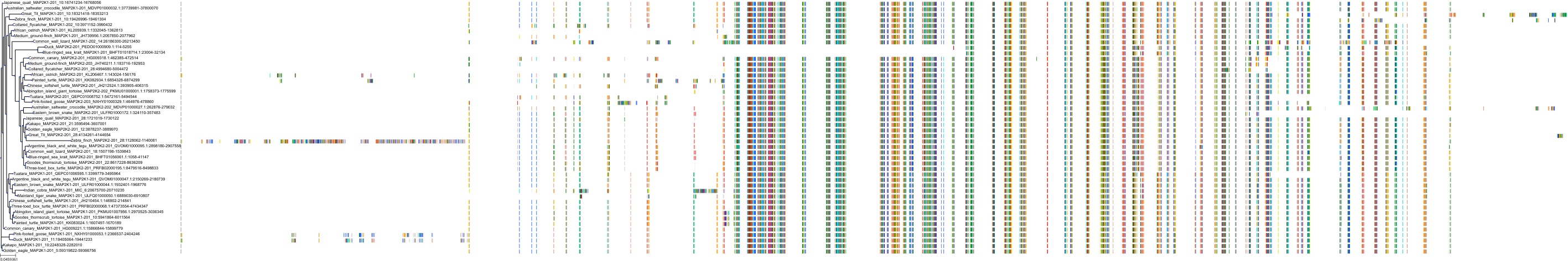
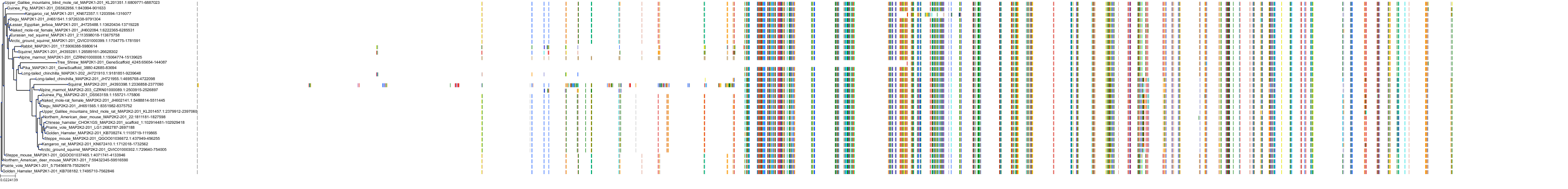
Protein: Dual specificity mitogen-activated protein kinase kinase 2 Description: Dual specificity mitogen-activated protein kinase kinase 2 Organism : Homo sapiens P36507 ENSG00000126934 |
||||
|
Protein: Dual specificity mitogen-activated protein kinase kinase 1 Description: Dual specificity mitogen-activated protein kinase kinase 1 Organism : Homo sapiens Q02750 ENSG00000169032 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEMBL | CHEMBL1236682 |
| DrugBank | DB06309 |
| FDA SRS | JPX07AFM0N |
| Guide to Pharmacology | 7942 |
| PDB | VRA |
| PubChem | 44182295 |
| SureChEMBL | SCHEMBL345333 |
| ZINC | ZINC000039187987 |