Structure
| InChI Key | MCIDWGZGWVSZMK-UHFFFAOYSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C26H28N6O |
| Molecular Weight | 440.55 |
| AlogP | 4.89 |
| Hydrogen Bond Acceptor | 5.0 |
| Hydrogen Bond Donor | 2.0 |
| Number of Rotational Bond | 5.0 |
| Polar Surface Area | 76.98 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 5.0 |
| Heavy Atoms | 33.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| PI3-kinase p110-delta subunit inhibitor | INHIBITOR | PubMed Other ClinicalTrials |
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Enzyme
Transferase
|
- | - | - | 0.1259 | - |
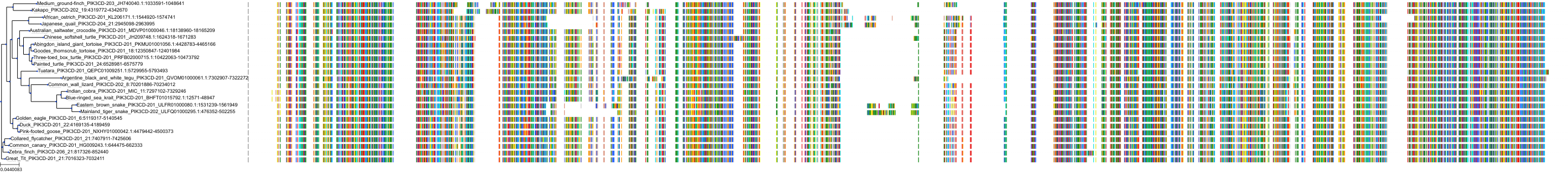
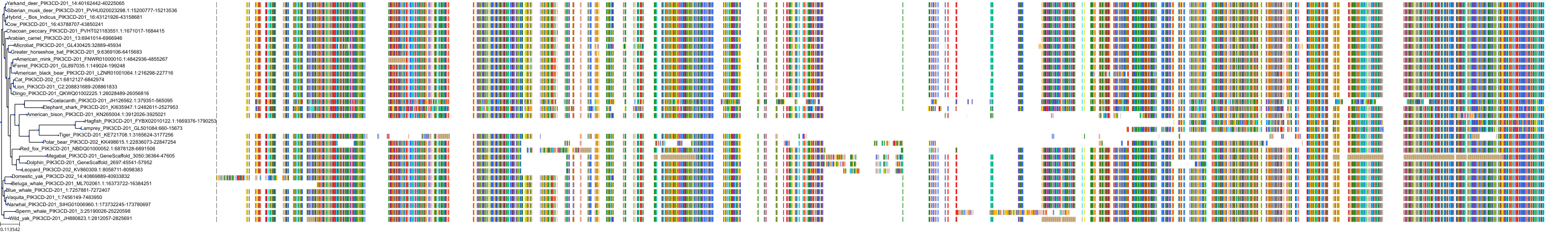
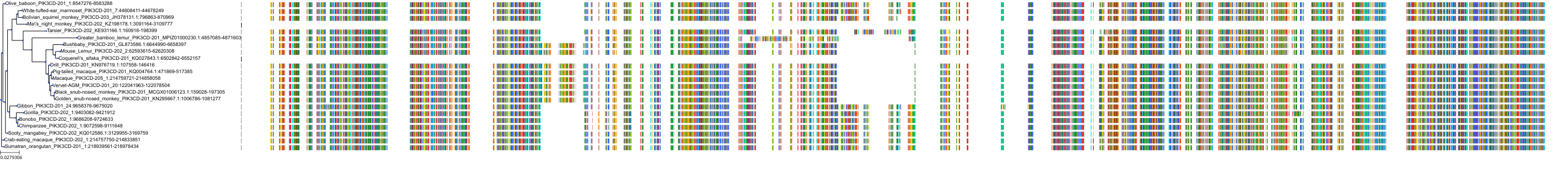
Target Conservation
|
Protein: PI3-kinase p110-delta subunit Description: Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform Organism : Homo sapiens O00329 ENSG00000171608 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEMBL | CHEMBL2216859 |
| DrugBank | DB16253 |
| FDA SRS | OEP8JJ3OZR |
| Guide to Pharmacology | 9425 |
| PDB | VVX |
| PubChem | 49784002 |
| SureChEMBL | SCHEMBL109919 |
| ZINC | ZINC000095564436 |
 Homo sapiens
Homo sapiens