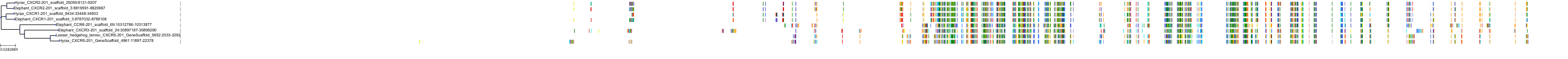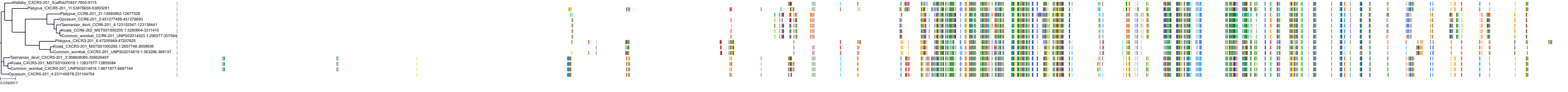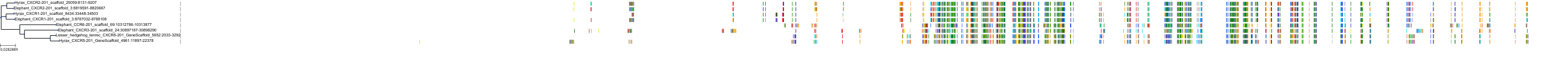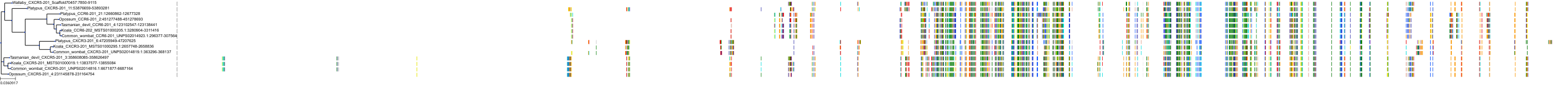Structure
| InChI Key | RXIUEIPPLAFSDF-CYBMUJFWSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C21H23N3O5 |
| Molecular Weight | 397.43 |
| AlogP | 2.9 |
| Hydrogen Bond Acceptor | 7.0 |
| Hydrogen Bond Donor | 3.0 |
| Number of Rotational Bond | 7.0 |
| Polar Surface Area | 111.88 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 3.0 |
| Heavy Atoms | 29.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Interleukin-8 receptor A antagonist | ANTAGONIST | PubMed PubMed ClinicalTrials PubMed |
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Membrane receptor
Family A G protein-coupled receptor
Peptide receptor (family A GPCR)
Chemokine receptor
CXC chemokine receptor
|
- | 0.049-680 | - | 4-20 | - |
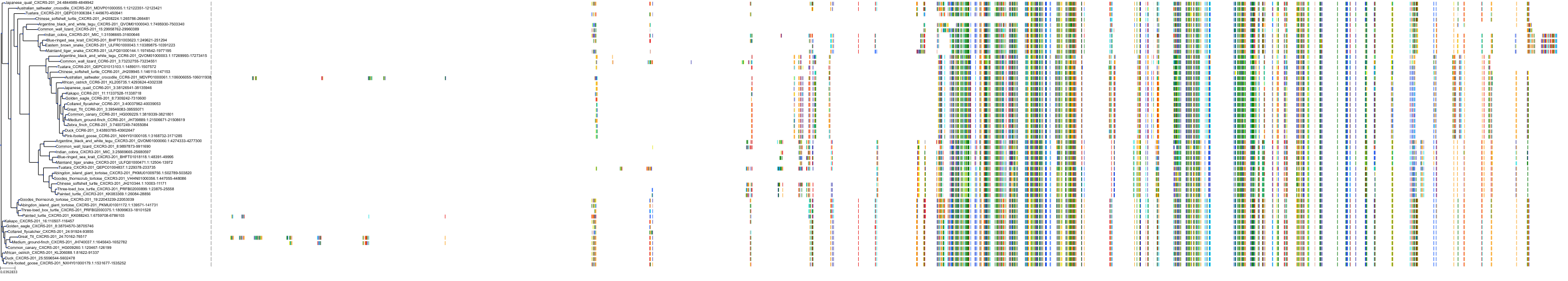
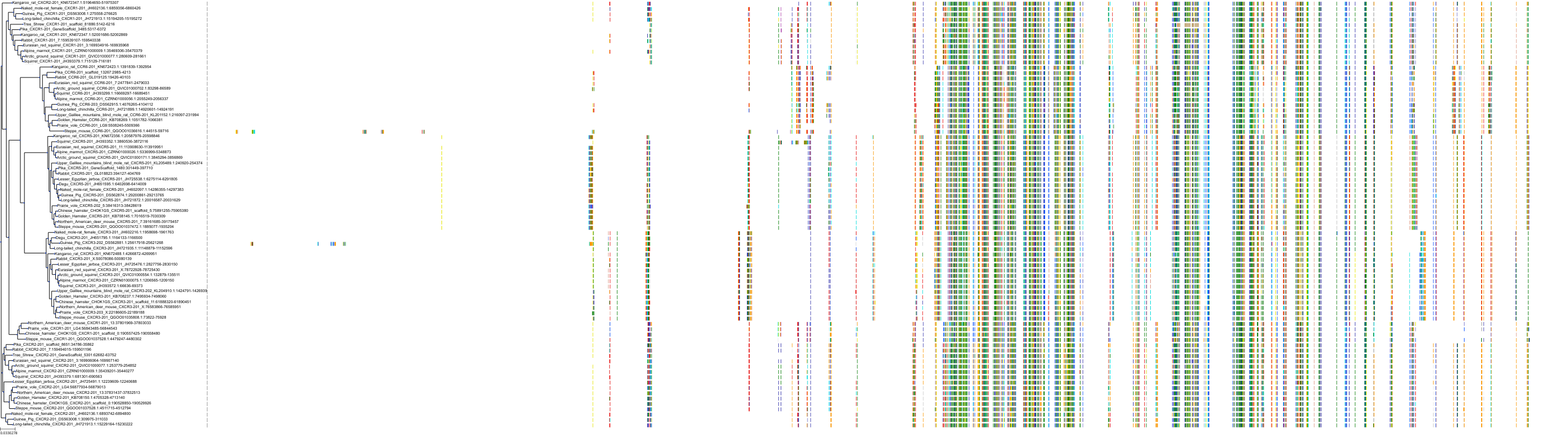
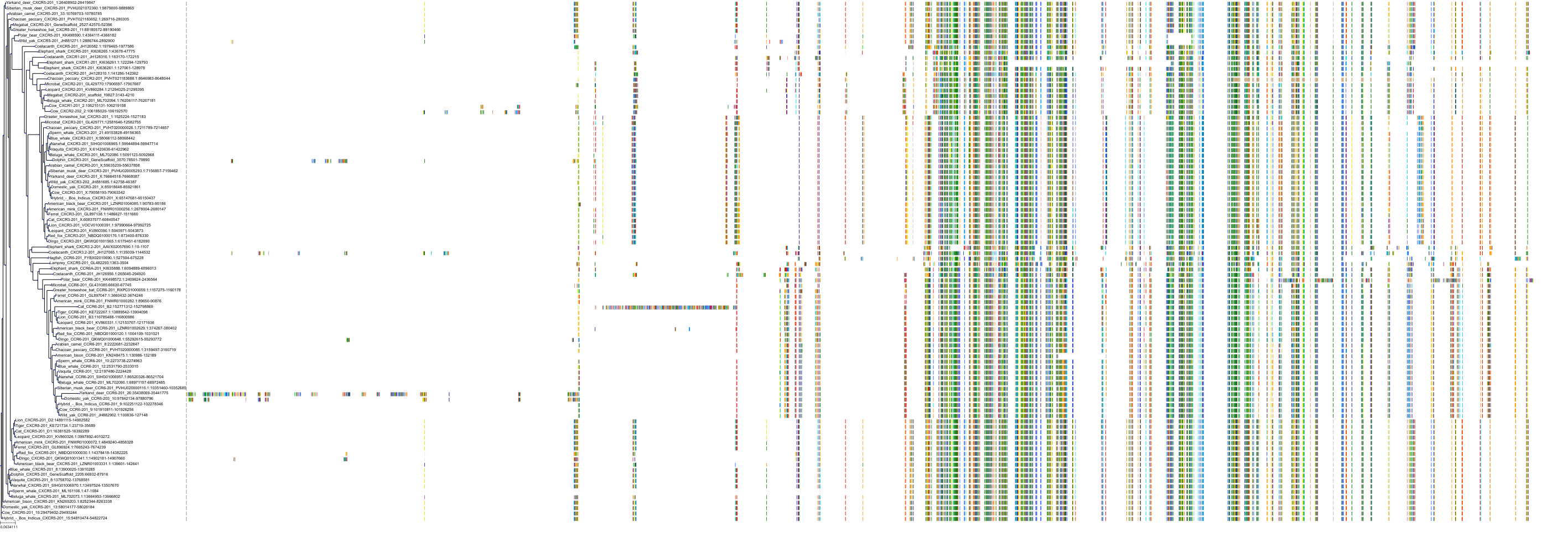
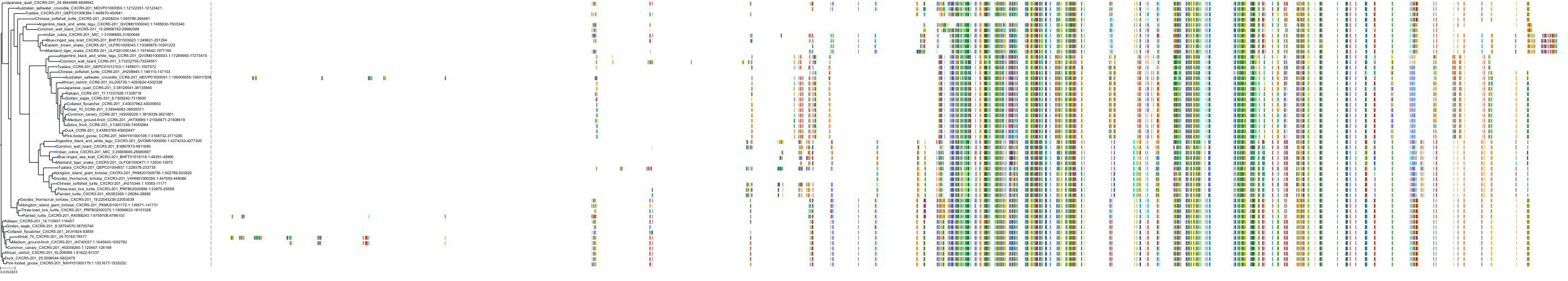
Target Conservation
|
Protein: Interleukin-8 receptor A Description: C-X-C chemokine receptor type 1 Organism : Homo sapiens P25024 ENSG00000163464 |
||||
|
Protein: Interleukin-8 receptor B Description: C-X-C chemokine receptor type 2 Organism : Homo sapiens P25025 ENSG00000180871 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEMBL | CHEMBL2103864 |
| FDA SRS | 7V3BY6G538 |
| Guide to Pharmacology | 8497 |
| PubChem | 71587828 |
| SureChEMBL | SCHEMBL2024684 |
| ChEMBL | CHEMBL216981 |
| FDA SRS | 7V3BY6G538 |
| Guide to Pharmacology | 8497 |
| PubChem | 71587828 |
| SureChEMBL | SCHEMBL184744 |
| ZINC | ZINC000100033051 |
 Homo sapiens
Homo sapiens
 Mus musculus
Mus musculus