| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| UNII | U1JK633AYI |
| EPA CompTox | DTXSID10196488 |
Structure
| InChI Key | RAHBGWKEPAQNFF-UHFFFAOYSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C22H23N5O |
| Molecular Weight | 373.46 |
| AlogP | 4.04 |
| Hydrogen Bond Acceptor | 5.0 |
| Hydrogen Bond Donor | 3.0 |
| Number of Rotational Bond | 5.0 |
| Polar Surface Area | 78.94 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 3.0 |
| Heavy Atoms | 28.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Platelet-derived growth factor receptor inhibitor | INHIBITOR | PubMed |
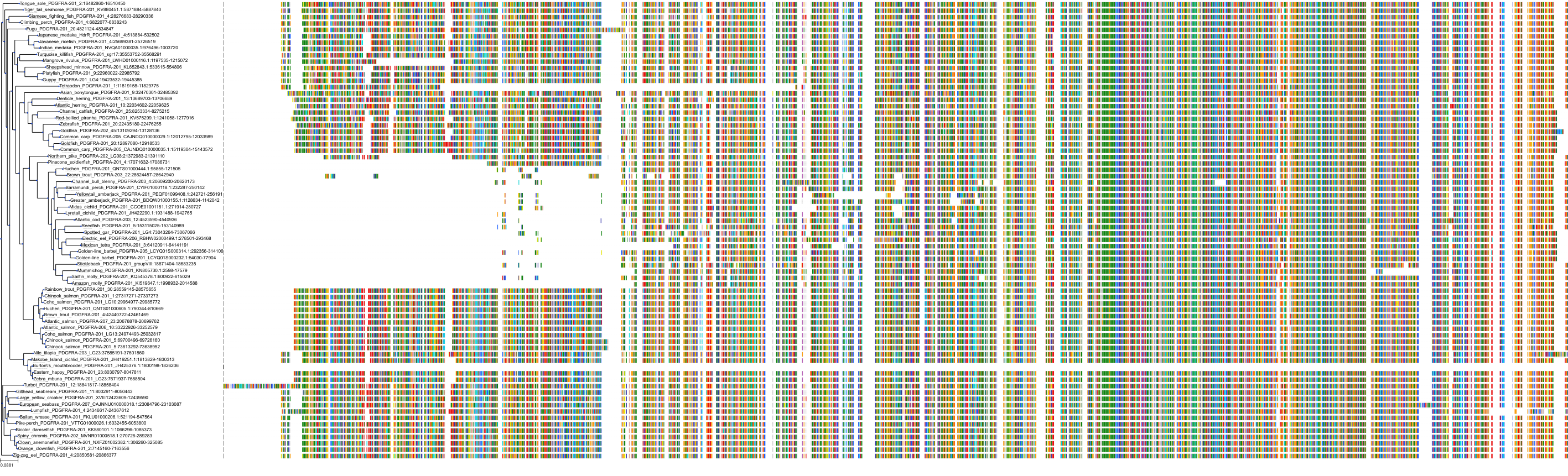
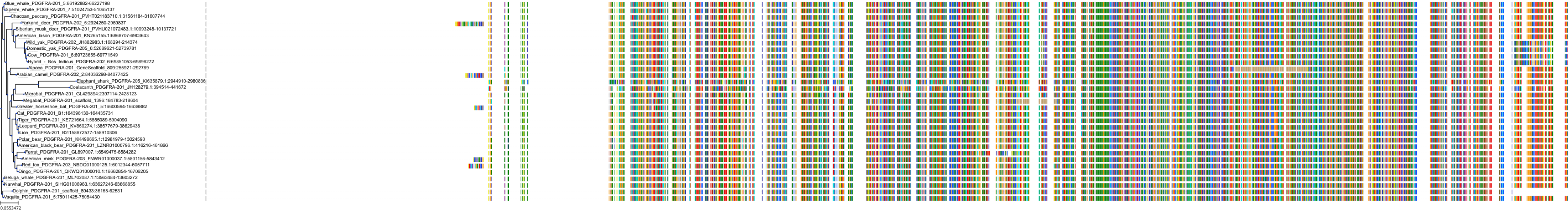
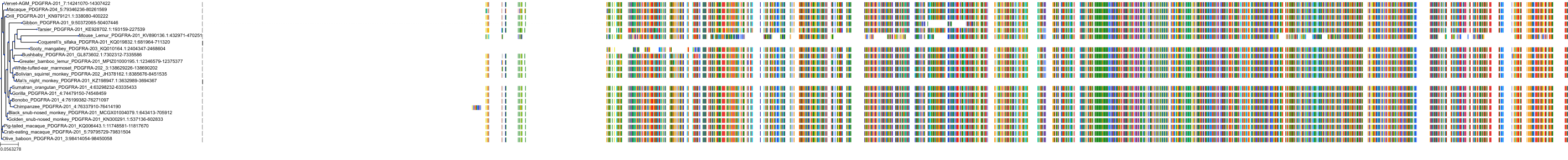
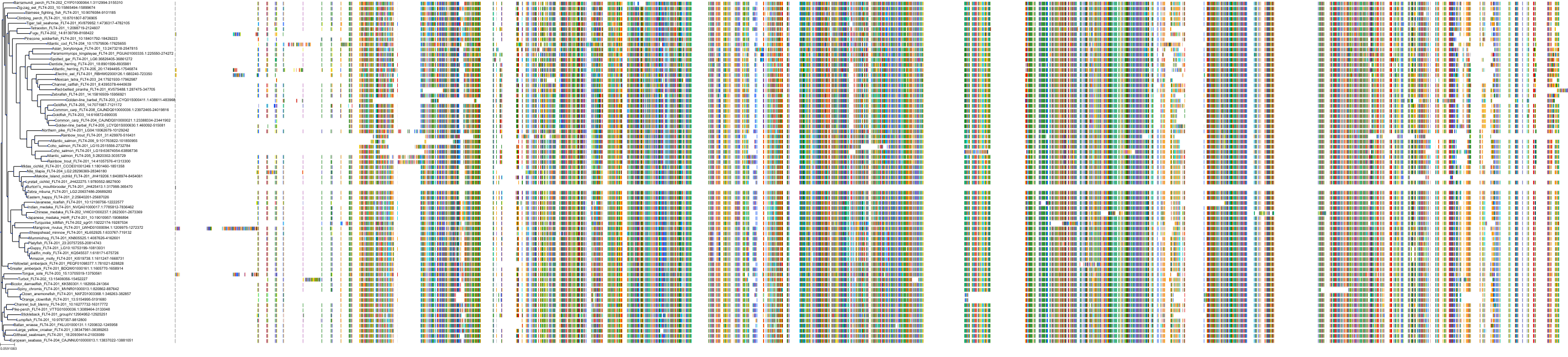
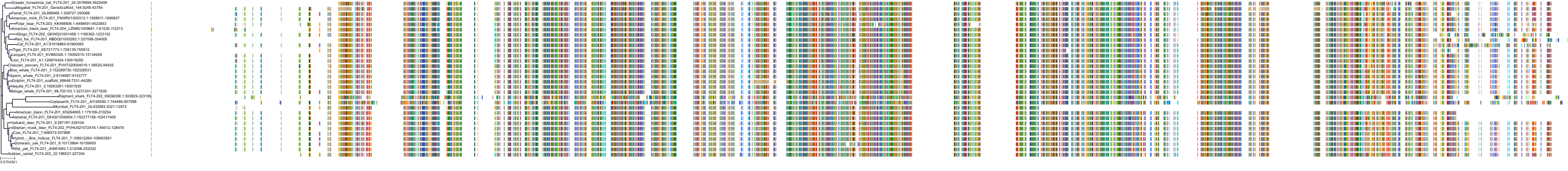
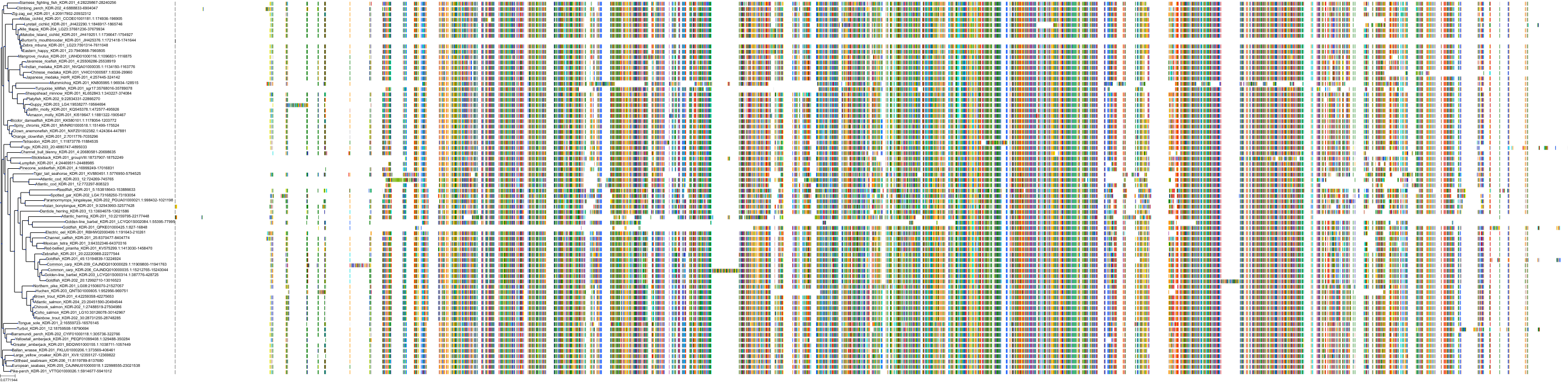
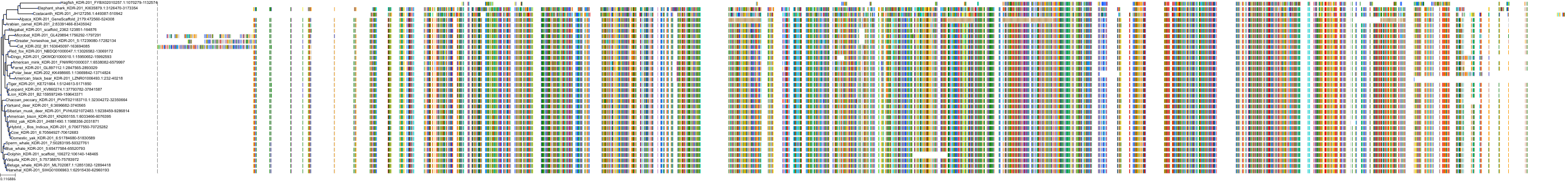
Target Conservation
|
Protein: Tyrosine-protein kinase receptor RET Description: Proto-oncogene tyrosine-protein kinase receptor Ret Organism : Homo sapiens P07949 ENSG00000165731 |
||||
|
Protein: Platelet-derived growth factor receptor Description: Platelet-derived growth factor receptor beta Organism : Homo sapiens P09619 ENSG00000113721 |
||||
|
Protein: Stem cell growth factor receptor Description: Mast/stem cell growth factor receptor Kit Organism : Homo sapiens P10721 ENSG00000157404 |
||||
|
Protein: Platelet-derived growth factor receptor Description: Platelet-derived growth factor receptor alpha Organism : Homo sapiens P16234 ENSG00000134853 |
||||
|
Protein: Vascular endothelial growth factor receptor Description: Vascular endothelial growth factor receptor 1 Organism : Homo sapiens P17948 ENSG00000102755 |
||||
|
Protein: Vascular endothelial growth factor receptor Description: Vascular endothelial growth factor receptor 3 Organism : Homo sapiens P35916 ENSG00000037280 |
||||
|
Protein: Vascular endothelial growth factor receptor Description: Vascular endothelial growth factor receptor 2 Organism : Homo sapiens P35968 ENSG00000128052 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 51098 |
| ChEMBL | CHEMBL572881 |
| DrugBank | DB05575 |
| FDA SRS | U1JK633AYI |
| Guide to Pharmacology | 5660 |
| PDB | 706 |
| PharmGKB | PA166118340 |
| PubChem | 11667893 |
| SureChEMBL | SCHEMBL187470 |
| ZINC | ZINC000018710082 |
 Homo sapiens
Homo sapiens