Structure
| InChI Key | KKYABQBFGDZVNQ-UHFFFAOYSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C22H26FN3O2 |
| Molecular Weight | 383.47 |
| AlogP | 3.86 |
| Hydrogen Bond Acceptor | 3.0 |
| Hydrogen Bond Donor | 2.0 |
| Number of Rotational Bond | 5.0 |
| Polar Surface Area | 71.09 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 2.0 |
| Heavy Atoms | 28.0 |
Pharmacology
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Enzyme
Kinase
Protein Kinase
CMGC protein kinase group
CMGC protein kinase MAPK family
CMGC protein kinase p38 subfamily
|
- | 25.12-100 | - | 7.943-63.1 | - |
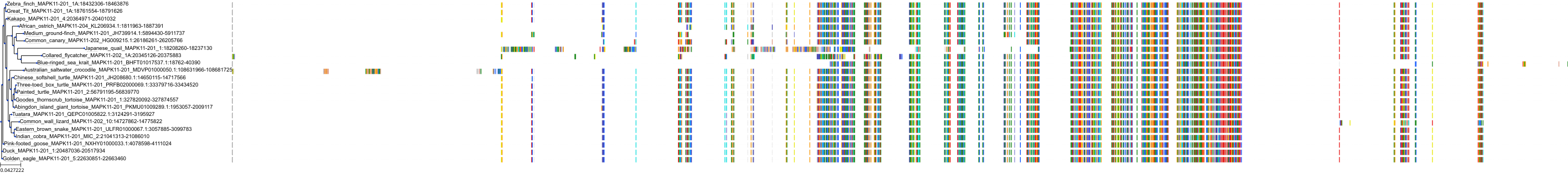
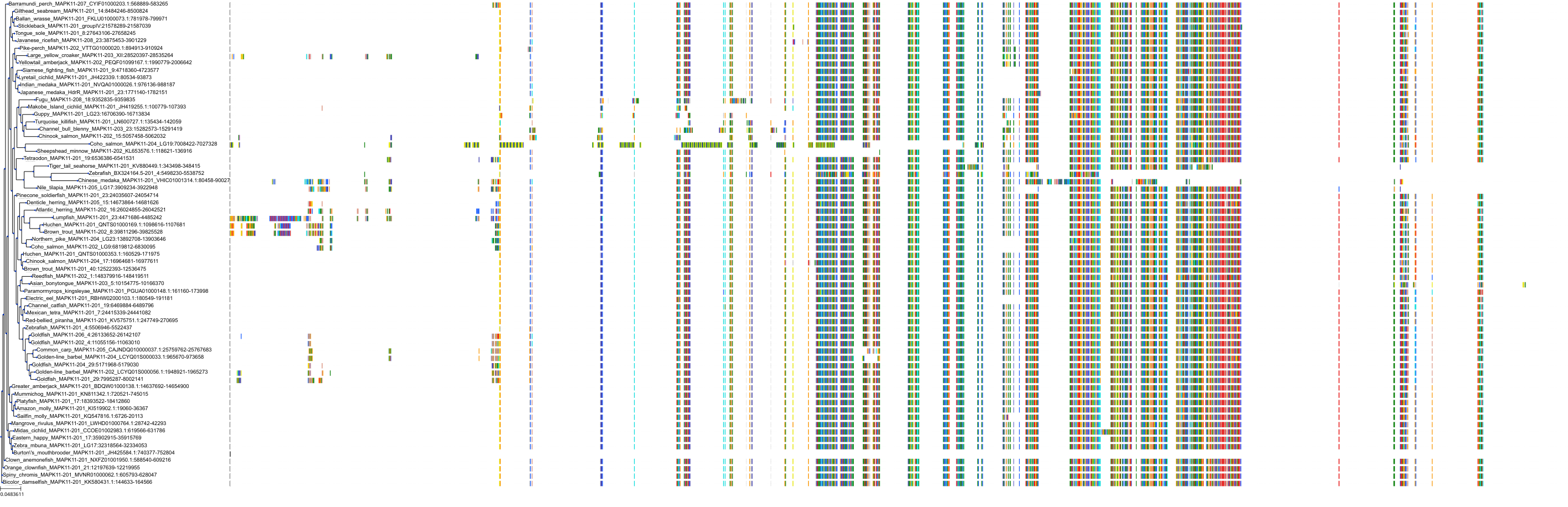
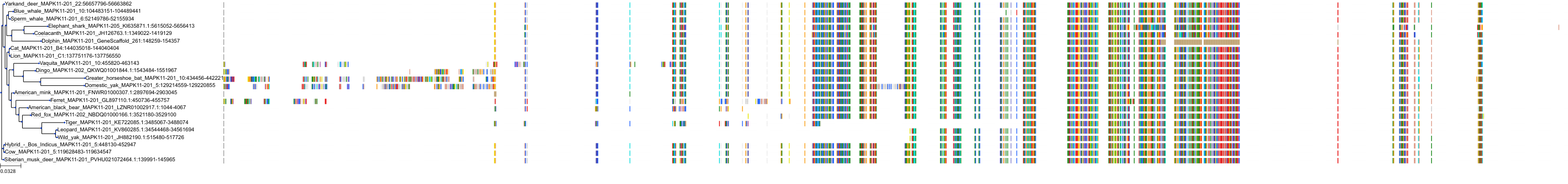
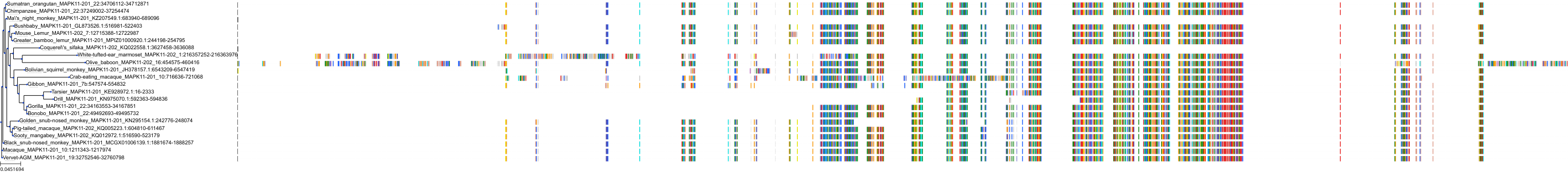
Target Conservation
|
Protein: MAP kinase p38 beta Description: Mitogen-activated protein kinase 11 Organism : Homo sapiens Q15759 ENSG00000185386 |
||||
|
Protein: MAP kinase p38 alpha Description: Mitogen-activated protein kinase 14 Organism : Homo sapiens Q16539 ENSG00000112062 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 131167 |
| ChEMBL | CHEMBL1088752 |
| DrugBank | DB12270 |
| FDA SRS | F2DQF16BXE |
| Guide to Pharmacology | 7835 |
| PubChem | 11552706 |
| SureChEMBL | SCHEMBL1070401 |
| ZINC | ZINC000035793138 |
 Homo sapiens
Homo sapiens