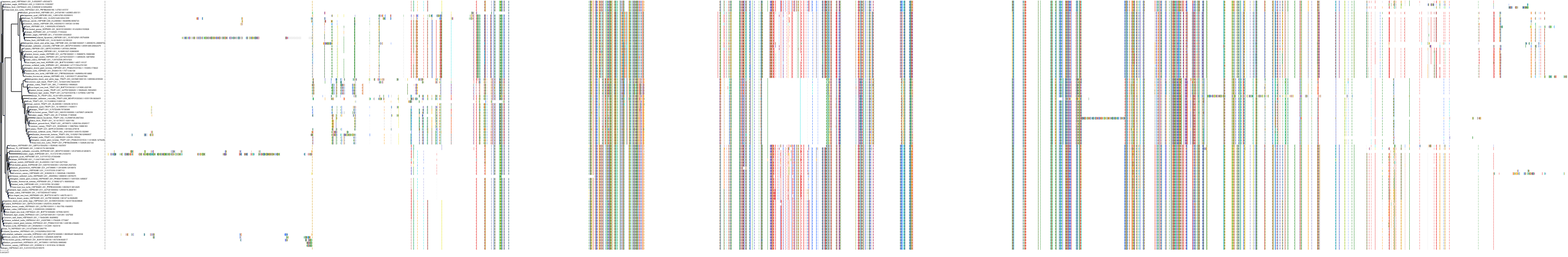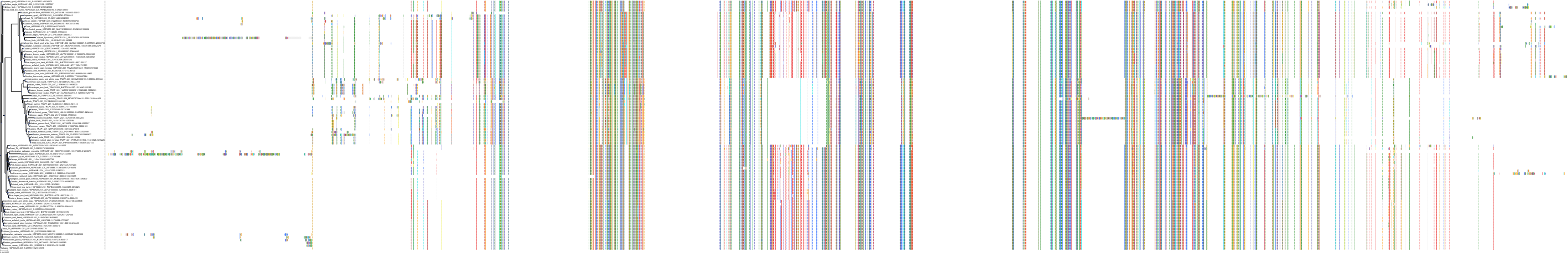Structure
| InChI Key | RVAQIUULWULRNW-UHFFFAOYSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C20H20N4O3 |
| Molecular Weight | 364.41 |
| AlogP | 3.25 |
| Hydrogen Bond Acceptor | 6.0 |
| Hydrogen Bond Donor | 3.0 |
| Number of Rotational Bond | 3.0 |
| Polar Surface Area | 96.07 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 4.0 |
| Heavy Atoms | 27.0 |
Pharmacology
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Other cytosolic protein
|
- | 5-63 | - | - | - | |
|
Other membrane protein
|
- | 10-37.7 | - | - | - |
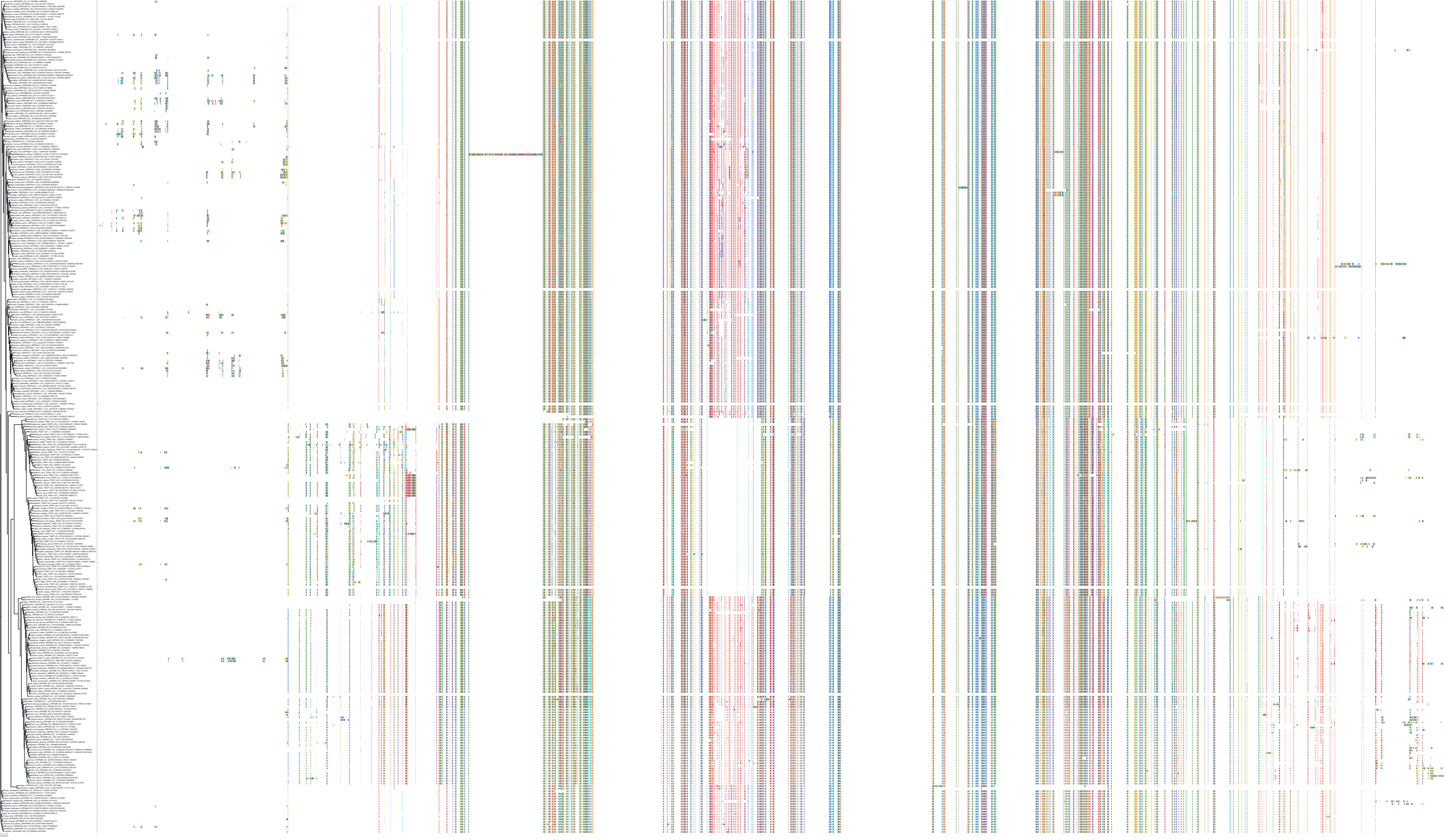
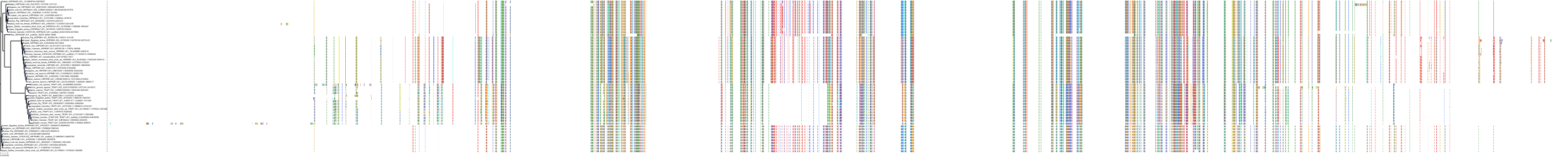
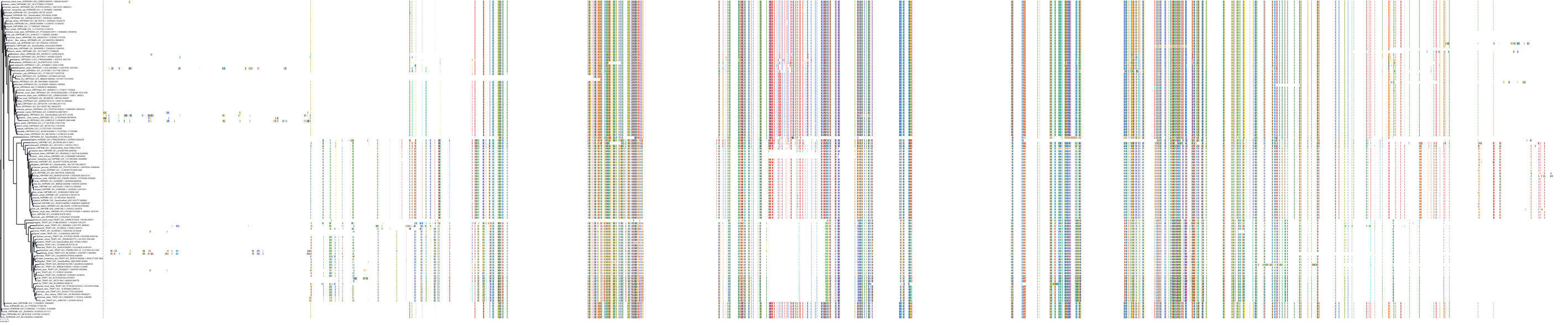
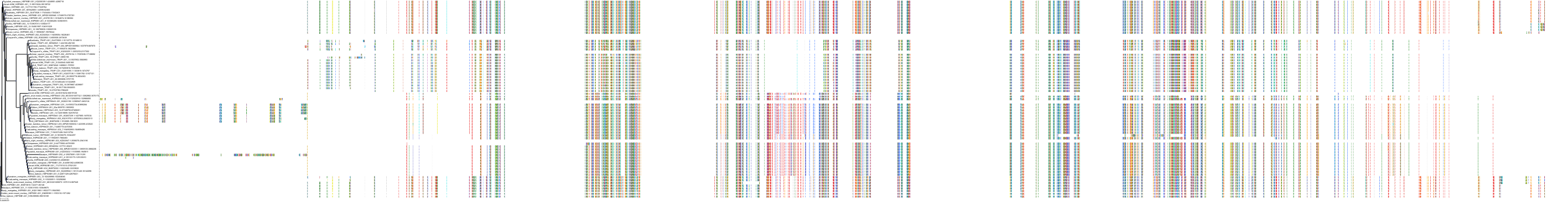
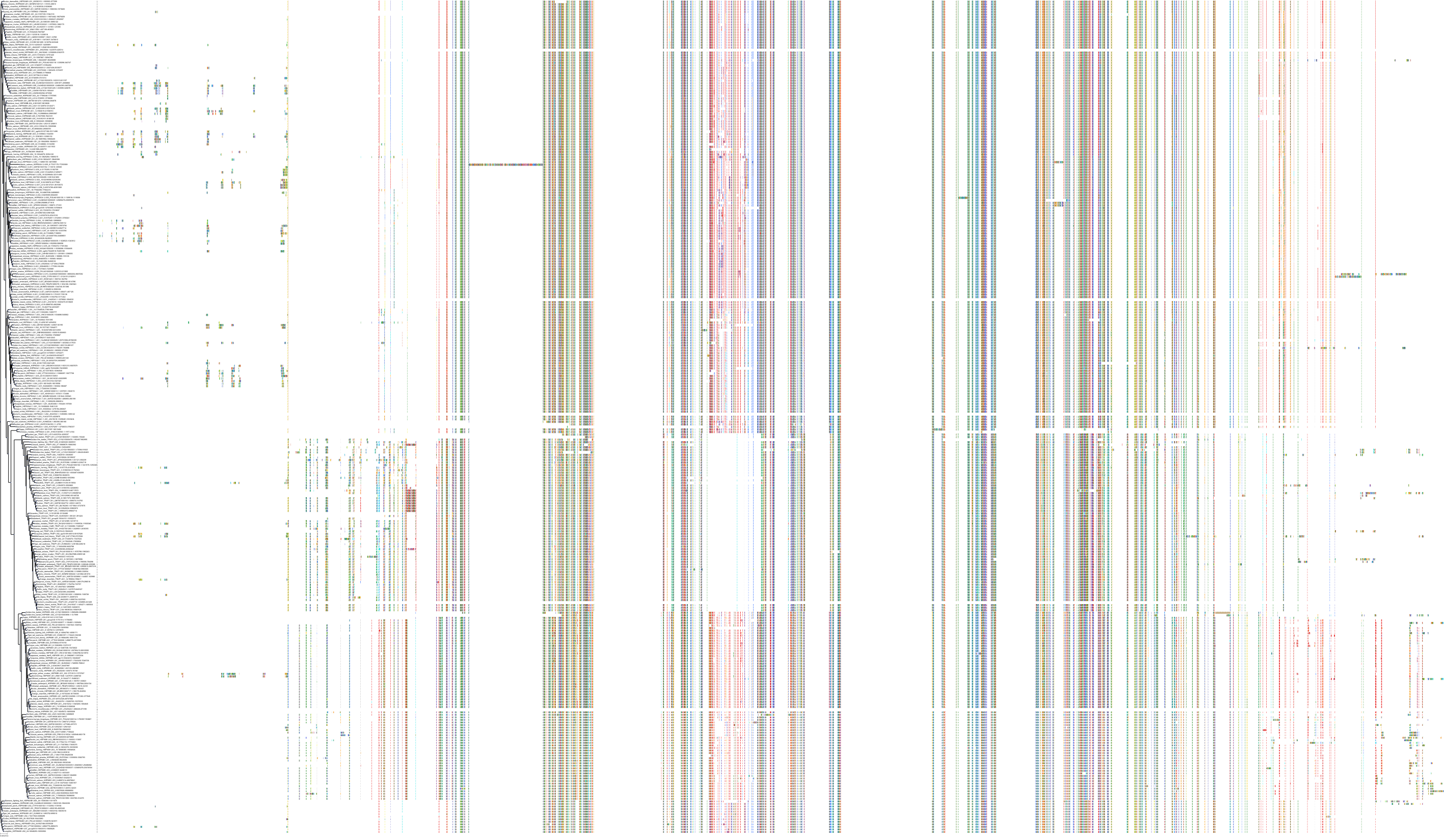
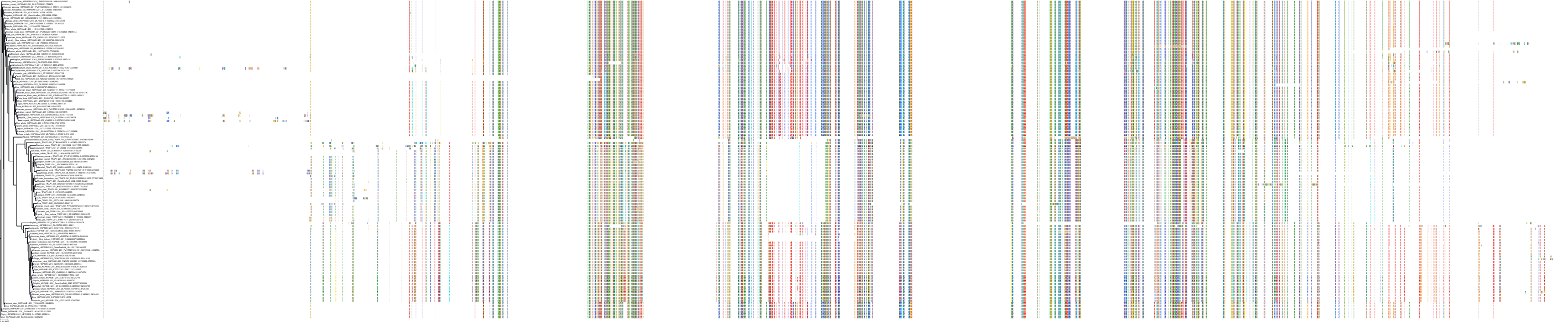
Target Conservation
|
Protein: Heat shock protein HSP90 Description: Heat shock protein HSP 90-alpha Organism : Homo sapiens P07900 ENSG00000080824 |
||||
|
Protein: Heat shock protein HSP90 Description: Heat shock protein HSP 90-beta Organism : Homo sapiens P08238 ENSG00000096384 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEMBL | CHEMBL2103879 |
| DrugBank | DB12047 |
| FDA SRS | 2E8412Y946 |
| PDB | TUH |
| PubChem | 135564985 |
| SureChEMBL | SCHEMBL419750 |
| ZINC | ZINC000043130413 |
 Canis lupus familiaris
Canis lupus familiaris
 Homo sapiens
Homo sapiens