| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| ATC | R06AX22 |
| UNII | TQD7Q784P1 |
| EPA CompTox | DTXSID6046472 |
Structure
| InChI Key | MJJALKDDGIKVBE-UHFFFAOYSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C32H39NO2 |
| Molecular Weight | 469.67 |
| AlogP | 7.22 |
| Hydrogen Bond Acceptor | 3.0 |
| Number of Rotational Bond | 9.0 |
| Polar Surface Area | 29.54 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 3.0 |
| Heavy Atoms | 35.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Histamine H1 receptor inverse agonist | INVERSE AGONIST | PubMed PubMed PubMed PubMed PubMed |
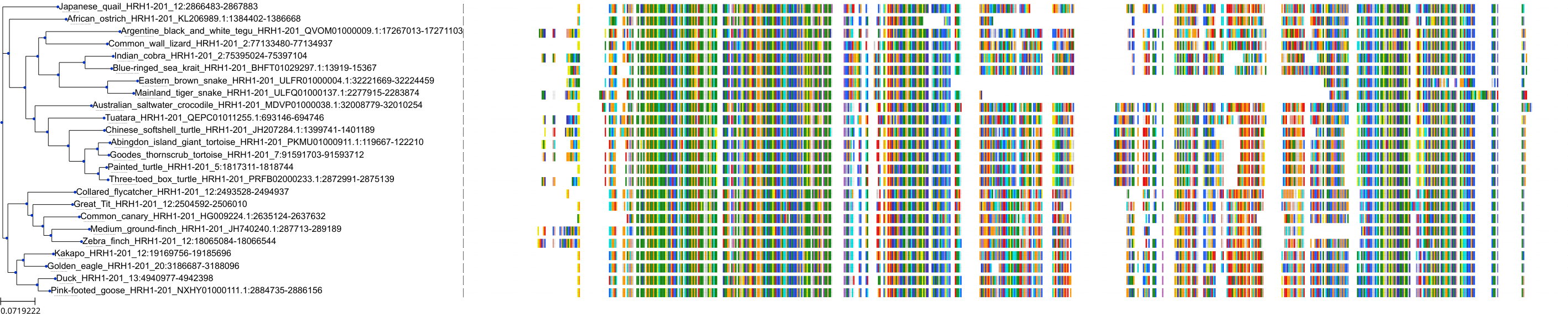
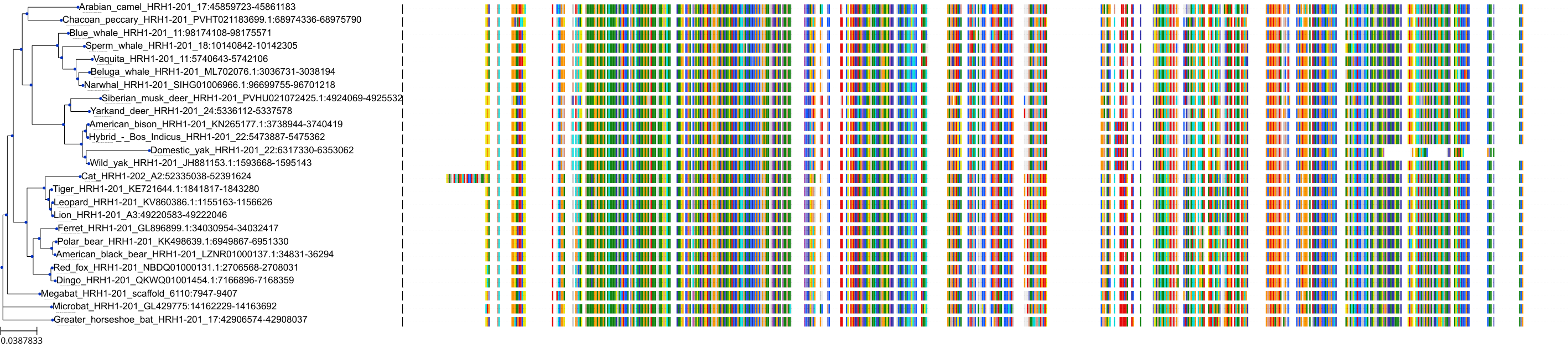
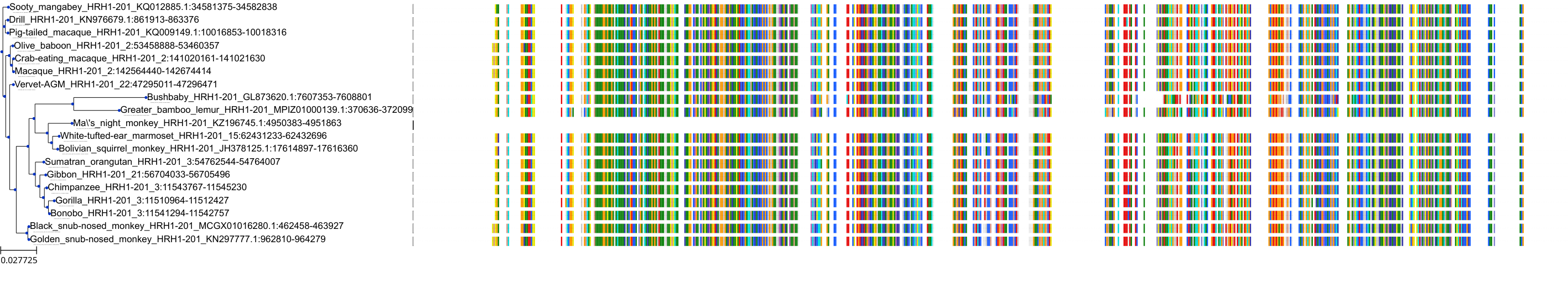
Target Conservation
|
Protein: Histamine H1 receptor Description: Histamine H1 receptor Organism : Homo sapiens P35367 ENSG00000196639 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 31528 |
| ChEMBL | CHEMBL305660 |
| DrugBank | DB11742 |
| DrugCentral | 977 |
| FDA SRS | TQD7Q784P1 |
| Human Metabolome Database | HMDB0060159 |
| PubChem | 3191 |
| SureChEMBL | SCHEMBL18467 |
| ZINC | ZINC000003781952 |
 Cavia porcellus
Cavia porcellus
 Homo sapiens
Homo sapiens
 Staphylococcus aureus
Staphylococcus aureus