| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| ATC | N07CA02 |
| UNII | 3DI2E1X18L |
| EPA CompTox | DTXSID3022821 |
Structure
| InChI Key | DERZBLKQOCDDDZ-JLHYYAGUSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C26H28N2 |
| Molecular Weight | 368.52 |
| AlogP | 5.11 |
| Hydrogen Bond Acceptor | 2.0 |
| Number of Rotational Bond | 6.0 |
| Polar Surface Area | 6.48 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 3.0 |
| Heavy Atoms | 28.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Histamine H1 receptor antagonist | ANTAGONIST | PubMed PubMed PubMed PubMed PubMed PubMed PubMed |
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Ion channel
Voltage-gated ion channel
Voltage-gated sodium channel
|
- | 440 | - | - | 94.6 | |
|
Transporter
Electrochemical transporter
SLC superfamily of solute carriers
SLC21/SLCO family of organic anion transporting polypeptides
|
- | - | - | - | 102.69-185.06 |
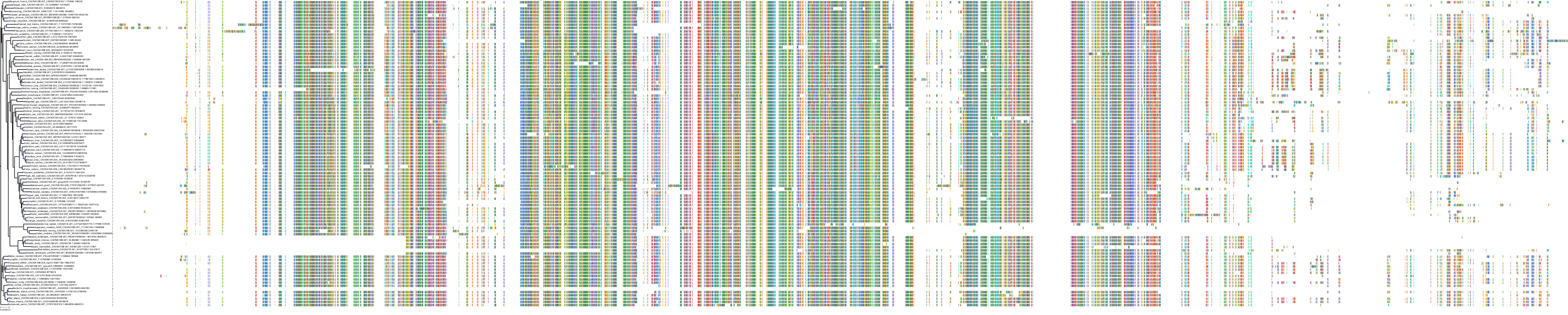
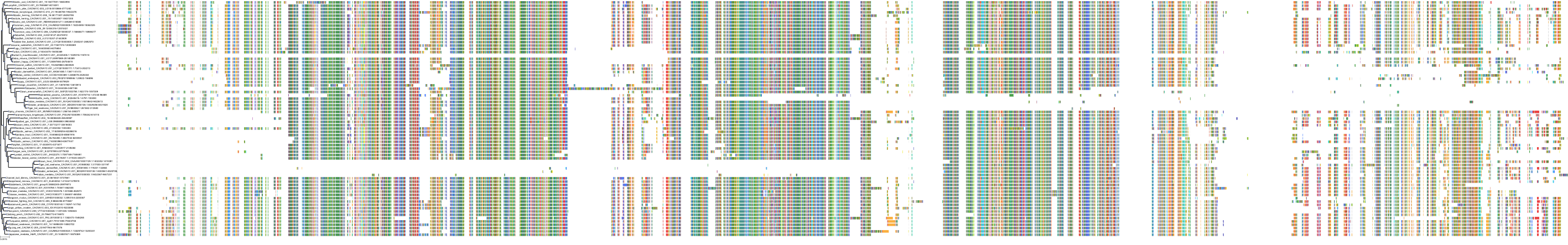
Target Conservation
|
Protein: Voltage-gated L-type calcium channel Description: Voltage-dependent L-type calcium channel subunit alpha-1F Organism : Homo sapiens O60840 ENSG00000102001 |
||||
|
Protein: Histamine H1 receptor Description: Histamine H1 receptor Organism : Homo sapiens P35367 ENSG00000196639 |
||||
|
Protein: Voltage-gated L-type calcium channel Description: Voltage-dependent L-type calcium channel subunit alpha-1D Organism : Homo sapiens Q01668 ENSG00000157388 |
||||
|
Protein: Voltage-gated L-type calcium channel Description: Voltage-dependent L-type calcium channel subunit alpha-1S Organism : Homo sapiens Q13698 ENSG00000081248 |
||||
|
Protein: Voltage-gated L-type calcium channel Description: Voltage-dependent L-type calcium channel subunit alpha-1C Organism : Homo sapiens Q13936 ENSG00000151067 |
||||
Related Entries
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 31403 |
| ChEMBL | CHEMBL43064 |
| DrugCentral | 654 |
| FDA SRS | 3DI2E1X18L |
| Guide to Pharmacology | 9072 |
| PubChem | 1547484 |
| SureChEMBL | SCHEMBL44957 |
| ZINC | ZINC000019632891 |
 Cavia porcellus
Cavia porcellus
 Cricetulus griseus
Cricetulus griseus