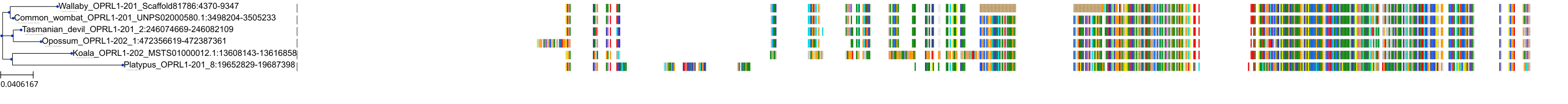Structure
| InChI Key | CSMVOZKEWSOFER-RQNOJGIXSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C24H27FN2O |
| Molecular Weight | 378.49 |
| AlogP | 5.11 |
| Hydrogen Bond Acceptor | 2.0 |
| Hydrogen Bond Donor | 1.0 |
| Number of Rotational Bond | 2.0 |
| Polar Surface Area | 28.26 |
| Molecular species | BASE |
| Aromatic Rings | 3.0 |
| Heavy Atoms | 28.0 |
Pharmacology
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Membrane receptor
Family A G protein-coupled receptor
Peptide receptor (family A GPCR)
Short peptide receptor (family A GPCR)
Opioid receptor
|
1.2-13 | - | - | 0.7-0.9 | - |
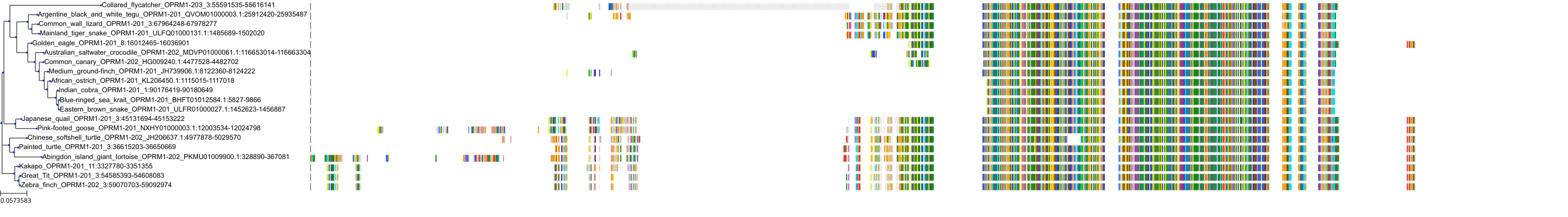
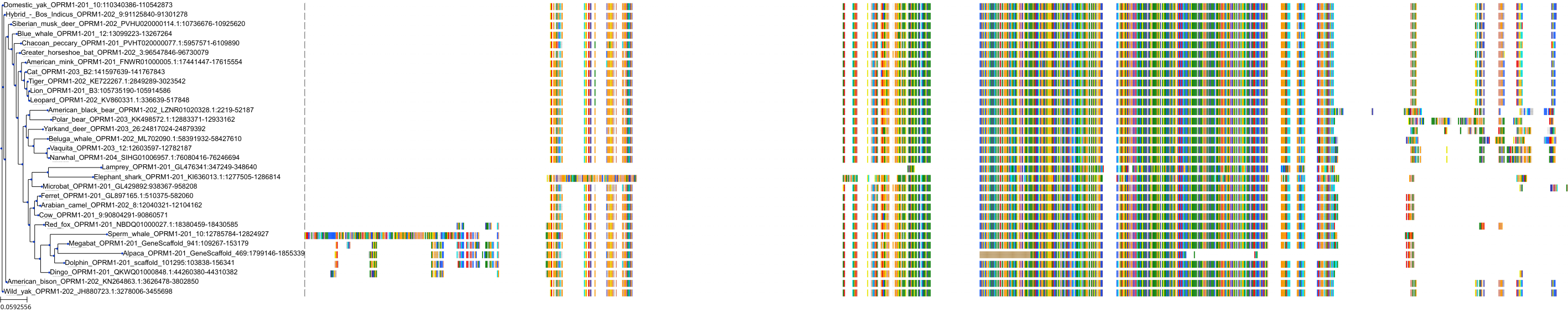
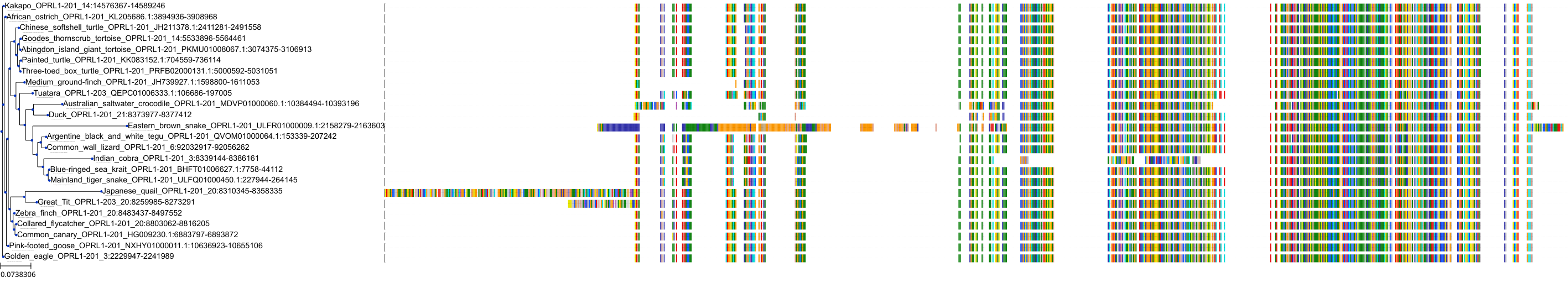
Target Conservation
|
Protein: Opioid receptors; mu/kappa/delta Description: Mu-type opioid receptor Organism : Homo sapiens P35372 ENSG00000112038 |
||||
|
Protein: Opioid receptors; mu/kappa/delta Description: Delta-type opioid receptor Organism : Homo sapiens P41143 ENSG00000116329 |
||||
|
Protein: Opioid receptors; mu/kappa/delta Description: Kappa-type opioid receptor Organism : Homo sapiens P41145 ENSG00000082556 |
||||
|
Protein: Nociceptin receptor Description: Nociceptin receptor Organism : Homo sapiens P41146 ENSG00000125510 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEMBL | CHEMBL2364605 |
| DrugBank | DB12830 |
| FDA SRS | 7GDW9S3GN3 |
| Guide to Pharmacology | 8866 |
| PubChem | 11848225 |
| SureChEMBL | SCHEMBL10035739 |
 Homo sapiens
Homo sapiens