Structure
| InChI Key | LDXYBEHACFJIEL-HNNXBMFYSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C19H25N7O2 |
| Molecular Weight | 383.46 |
| AlogP | 0.65 |
| Hydrogen Bond Acceptor | 7.0 |
| Hydrogen Bond Donor | 2.0 |
| Number of Rotational Bond | 6.0 |
| Polar Surface Area | 115.42 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 2.0 |
| Heavy Atoms | 28.0 |
Pharmacology
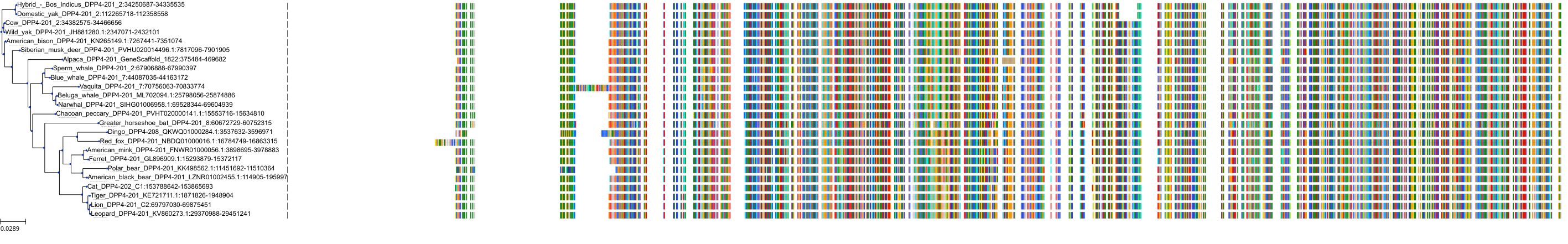
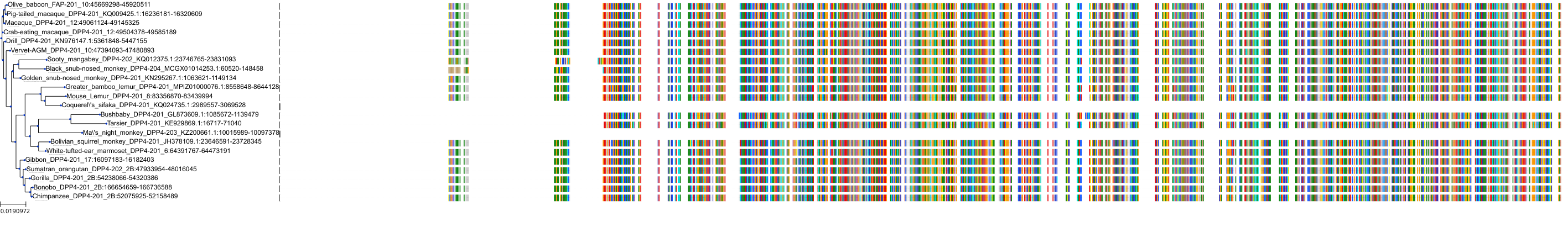
Target Conservation
|
Protein: Dipeptidyl peptidase IV Description: Dipeptidyl peptidase 4 Organism : Homo sapiens P27487 ENSG00000197635 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 136043 |
| ChEMBL | CHEMBL1929396 |
| DrugBank | DB12417 |
| DrugCentral | 4896 |
| FDA SRS | K726J96838 |
| PDB | SKK |
| PubChem | 44513473 |
| SureChEMBL | SCHEMBL905393 |
| ZINC | ZINC000070647144 |