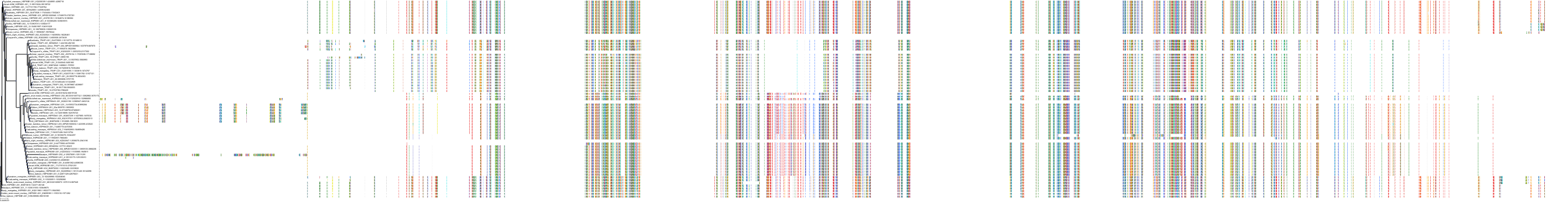
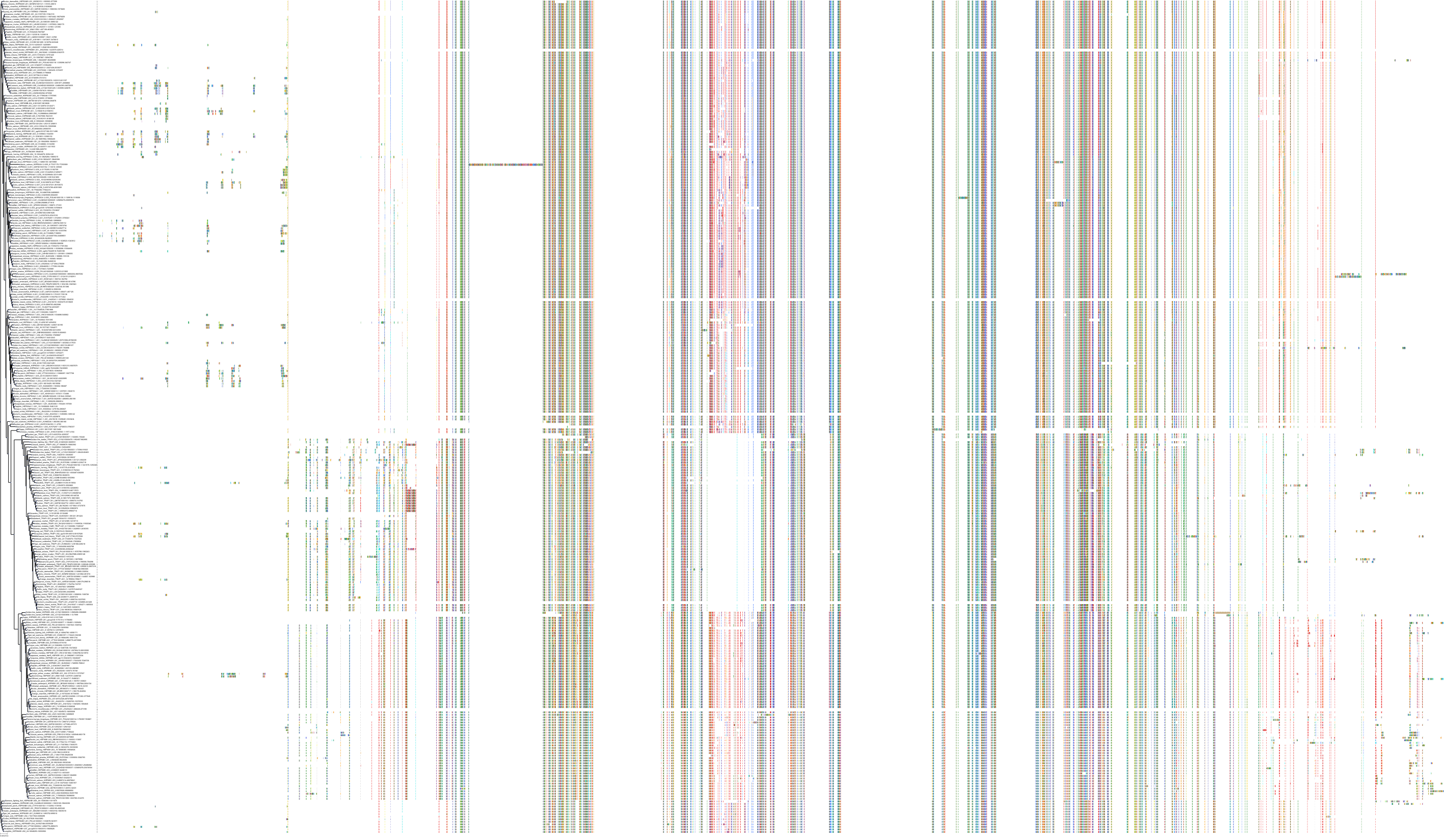
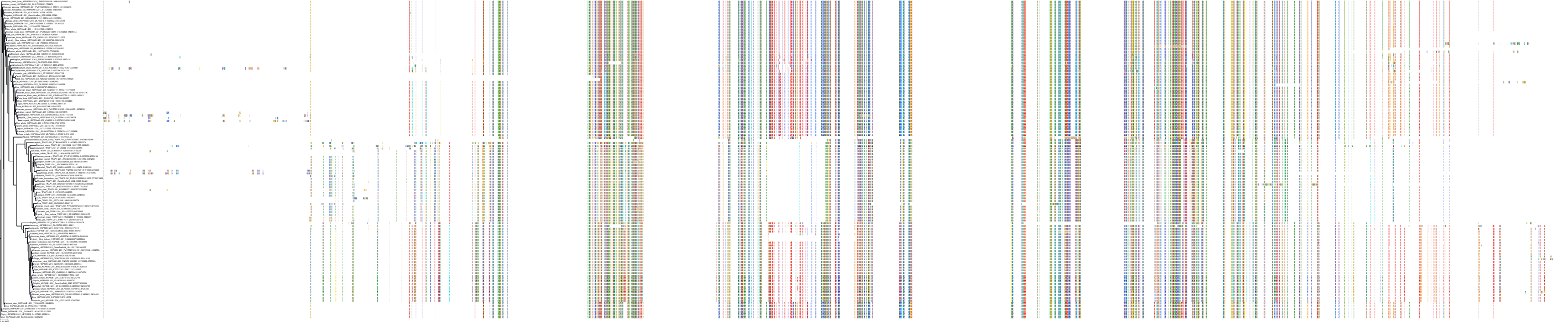
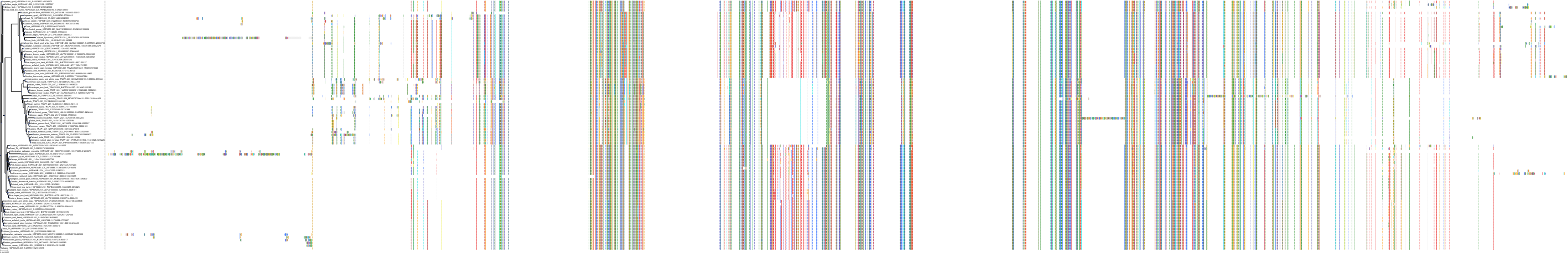|
Binding affinity for human heat shock protein 90 in scintillation proximity assay
|
Homo sapiens
|
500.0
nM
|
|
|
Cytotoxicity against SKBr3 cells
|
Homo sapiens
|
24.0
nM
|
|
|
Inhibition of BODIPY-AG binding to human HSP90
|
Homo sapiens
|
62.0
nM
|
|
|
Inhibition of BODIPY-AG binding to dog Grp94
|
Canis lupus familiaris
|
65.0
nM
|
|
|
Degradation of Her2 in SKBR3 cells
|
Homo sapiens
|
8.0
nM
|
|
|
Degradation of Her2 in SKOV3 cells
|
Homo sapiens
|
46.0
nM
|
|
|
Upregulation of Hsp70 in SKBR3 cells
|
Homo sapiens
|
4.0
nM
|
|
|
Upregulation of Hsp70 in SKOV3 cells
|
Homo sapiens
|
14.0
nM
|
|
|
Inhibition of HIF1 activation in human AGS cells assessed as inhibition of hypoxia-induced luciferase expression after 16 hrs by reporter assay
|
Homo sapiens
|
36.0
nM
|
|
|
Inhibition of HIF1 activation in human Hep3B cells assessed as inhibition of hypoxia-induced luciferase expression after 16 hrs by reporter assay
|
Homo sapiens
|
61.0
nM
|
|
|
Inhibition of hypoxia-induced HIF1 activation in human AGS cells by reporter gene assay
|
Homo sapiens
|
3.6
nM
|
|
|
Inhibition of TNF-alpha-induced NF-kappaB activation in human HeLa cells
|
Homo sapiens
|
150.0
nM
|
|
|
Inhibition of Hsp90 in human MDA-MB-231 cells assessed as Akt degradation
|
Homo sapiens
|
17.6
nM
|
|
|
Inhibition of Hsp90 in human MDA-MB-231 cells assessed as her2 degradation
|
Homo sapiens
|
4.5
nM
|
|
|
Cytotoxicity against human MDA-MB-231 cells by MTT assay
|
Homo sapiens
|
5.8
nM
|
|
|
Inhibition of Hsp90 in human A2058 cells assessed as Akt degradation
|
Homo sapiens
|
24.3
nM
|
|
|
Cytotoxicity against human A2058 cells by MTT assay
|
Homo sapiens
|
2.1
nM
|
|
|
Inhibition of Hsp90 in human A2058 cells
|
Homo sapiens
|
7.9
nM
|
|
|
Cytotoxicity against human SKBR3 cells after 72 hrs by celltiter-glo assay
|
Homo sapiens
|
24.0
nM
|
|
|
Binding affinity to human recombinant HSP90
|
Homo sapiens
|
500.0
nM
|
|
|
Cytotoxicity against human MCF7 cells after 72 hrs by celltiter-glo assay
|
Homo sapiens
|
230.0
nM
|
|
|
Cytotoxicity against human SKOV3 cells after 72 hrs by celltiter-glo assay
|
Homo sapiens
|
220.0
nM
|
|
|
Cytotoxicity against human A549 cells after 72 hrs by celltiter-glo assay
|
Homo sapiens
|
68.0
nM
|
|
|
Cytotoxicity against human CCRF-CEM cells after 72 hrs by celltiter-96 aqueous one solution assay
|
Homo sapiens
|
540.0
nM
|
|
|
Cytotoxicity against human MCF7 cells after 72 hrs in presence of NQO1 inhibitor dicoumarol
|
Homo sapiens
|
862.0
nM
|
|
|
Cytotoxicity against human SKBR3 cells after 72 hrs in presence of NQO1 inhibitor dicoumarol
|
Homo sapiens
|
230.0
nM
|
|
|
Binding affinity to human recombinant Hsp90alpha N-terminal domain by scintillation proximity assay
|
Homo sapiens
|
500.0
nM
|
|
|
Binding affinity to human recombinant Hsp90alpha N-terminal domain by isothermal titration calorimetry
|
Homo sapiens
|
87.0
nM
|
|
|
Cytotoxicity against human SKBR3 cells after 72 hrs
|
Homo sapiens
|
58.0
nM
|
|
|
Cytotoxicity against human SKOV3 cells after 72 hrs
|
Homo sapiens
|
122.0
nM
|
|
|
Cytotoxicity against human HCT116 cells after 72 hrs
|
Homo sapiens
|
57.0
nM
|
|
|
Cytotoxicity against human MCF7 cells after 72 hrs
|
Homo sapiens
|
71.0
nM
|
|
|
Binding affinity to Hsp90 in human SKBR3 cells
|
Homo sapiens
|
24.0
nM
|
|
|
Inhibition of hypoxia-induced HIF1alpha protein accumulation in human Hep3B cells treated for 30 mins measured after 12 hrs by Western blot analysis
|
Homo sapiens
|
57.2
nM
|
|
|
Inhibition of hypoxia-induced VEGF protein secretion in human Hep3B cells after 16 hrs by ELISA
|
Homo sapiens
|
79.5
nM
|
|
|
Displacement of [3H]pGA from His-tagged Hsp90 by scintillation proximity assay
|
None
|
250.0
nM
|
|
|
Inhibition of Hsp90 in human H1299 assessed as degradation of Hsp90 client protein Akt
|
Homo sapiens
|
100.0
nM
|
|
|
Inhibition of hypoxia-induced HIF1alpha protein accumulation in human Hep3B cells treated for 30 mins measured after 12 hrs by Western blot analysis
|
Homo sapiens
|
57.0
nM
|
|
|
Inhibition of hypoxia-induced VEGF protein secretion in human Hep3B cells after 16 hrs by ELISA
|
Homo sapiens
|
79.0
nM
|
|
|
Binding affinity to Hsp90 N-terminal ATPase domain by isothermal titration calorimetry assay
|
None
|
210.0
nM
|
|
|
Cytotoxicity against human HCT116 cells by Alamar blue assay
|
Homo sapiens
|
50.0
nM
|
|
|
Antiviral activity against Hepatitis C virus genotype 1b Con1 infected in human HuH7 cells assessed as GAPDH RNA or 18S rRNA level after 3 days selected with 40 nM HCV-796 and 800 nM boceprevir by qRT-PCR analysis
|
Hepatitis C virus
|
3.1
nM
|
|
|
Antiviral activity against Hepatitis C virus genotype 1b Con1 infected in human HuH7 cells assessed as GAPDH RNA or 18S rRNA level after 3 days by qRT-PCR analysis
|
Hepatitis C virus
|
1.2
nM
|
|
|
Inhibition of human HSP90 in human NCI-H1299 cells assessed as Akt degradation after 24 hrs by luminex assay
|
Homo sapiens
|
100.0
nM
|
|
|
Displacement of [3H]-(R)-2-(5-chloro-2,4-dihydroxybenzoyl)-N-ethylisoindoline-1-carboxamide from human his(6)-tagged HSP90alpha after 30 mins by scintillation proximity assay
|
Homo sapiens
|
680.0
nM
|
|
|
Inhibition of Hsp90 in human COLO205 cells xenografted in mouse assessed as Raf1 degradation at 100 mg/kg administered QD for 2 days measured 8 hrs post-last dose
|
Mus musculus
|
84.0
%
|
|
|
Antiproliferative activity against human HCT116 cells assessed as inhibition of cell viability after 48 hrs by MTT assay
|
Homo sapiens
|
780.0
nM
|
|
|
Antiproliferative activity against human MCF7 cells assessed as inhibition of cell viability after 48 hrs by MTT assay
|
Homo sapiens
|
390.0
nM
|
|
|
Inhibition of full-length Hsp90-ATPase activity (unknown origin) assessed as inhibition of ATP hydrolysis by spectrophotometry
|
Homo sapiens
|
930.0
nM
|
|
|
Inhibition of HSP90 (unknown origin)-mediated ATPase activity at 40 uM after 3 hrs by malachite green assay relative to control
|
Homo sapiens
|
88.31
%
|
|
|
Displacement of FITC-geldanamycin from HSP90 (unknown origin) after 30 mins by fluorescence polarization assay
|
Homo sapiens
|
90.0
nM
|
|
|
Antiproliferative activity against human MCF7 cells after 48 hrs by MTT assay
|
Homo sapiens
|
800.0
nM
|
|
|
Antiproliferative activity against human A231 cells after 48 hrs by MTT assay
|
Homo sapiens
|
170.0
nM
|
|
|
Drug metabolism in human liver microsomes assessed as 1 uM CYP3A inhibitor ketoconazole-mediated inhibition of (4E,6Z,8S,9S,10E,12S,13R,14S,16R)-19-(2-(dimethylamino)ethylamino)-13-hydroxy-10-(hydroxymethyl)-8,14-dimethoxy-4,12,16-trimethyl-3,20,22-trioxo-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate formation at 1 uM 17-DMAG by liquid chromatography-tandem mass spectrometry
|
Homo sapiens
|
75.0
%
|
|
|
Drug metabolism in human liver microsomes assessed as 1 uM CYP3A inhibitor ketoconazole-mediated inhibition of (4E,6Z,8S,9S,10E,12S,13R,14S,16R)-19-(2-(dimethylamino)-1-hydroxyethylamino)-13-hydroxy-8,14-dimethoxy-4,10,12,16-tetramethyl-3,20,22-trioxo-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate formation at 1 uM 17-DMAG by liquid chromatography-tandem mass spectrometry
|
Homo sapiens
|
77.0
%
|
|
|
Drug metabolism in human liver microsomes assessed as 1 uM CYP3A inhibitor ketoconazole-mediated inhibition of (4E,6Z,8S,9S,10E,12S,13R,14S,16R)-19-(2-(dimethylamino)ethylamino)-13,14-dihydroxy-8-methoxy-4,10,12,16-tetramethyl-3,20,22-trioxo-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate formation at 1 uM 17-DMAG by liquid chromatography-tandem mass spectrometry
|
Homo sapiens
|
79.0
%
|
|
|
Drug metabolism in human liver microsomes assessed as 1 uM CYP3A inhibitor ketoconazole-mediated inhibition of (4E,6Z,8S,9S,10E,12S,13R,14S,16R)-19-(2-(dimethylamino)ethylamino)-8,13-dihydroxy-14-methoxy-4,10,12,16-tetramethyl-3,20,22-trioxo-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate formation at 1 uM 17-DMAG by liquid chromatography-tandem mass spectrometry
|
Homo sapiens
|
81.0
%
|
|
|
Drug metabolism in human liver microsomes assessed as 1 uM CYP3A inhibitor ketoconazole-mediated inhibition of (4E,6Z,8S,9S,10E,12R,13R,14S,16R)-19-(2-(dimethylamino)-1-hydroxyethylamino)-13-hydroxy-12,16-bis(hydroxymethyl)-8,14-dimethoxy-4,10-dimethyl-3,20,22-trioxo-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate formation at 1 uM 17-DMAG by liquid chromatography-tandem mass spectrometry
|
Homo sapiens
|
83.0
%
|
|
|
Antiviral activity determined as inhibition of SARS-CoV-2 induced cytotoxicity of Caco-2 cells at 10 uM after 48 hours by high content imaging
|
Homo sapiens
|
-0.7
%
|
|
|
Inhibition of Hsp90 in human SKBR3 cells
|
Homo sapiens
|
24.0
nM
|
|
|
Cytotoxicity against HGF-induced erlotinib-resistant human PC9 cells assessed as inhibition of cell growth after 72 hrs by MTT assay
|
Homo sapiens
|
10.0
nM
|
|
|
Cytotoxicity against HGF-induced erlotinib-resistant human Ma1 cells assessed as inhibition of cell growth after 72 hrs by MTT assay
|
Homo sapiens
|
10.0
nM
|
|
|
Inhibition of 6x-His tagged human recombinant full length Hsp90alpha ATPase preincubated for 0.5 hrs followed by ATP addition measured after 30 mins by HTRF assay
|
Homo sapiens
|
919.0
nM
|
|
|
SARS-CoV-2 3CL-Pro protease inhibition percentage at 20µM by FRET kind of response from peptide substrate
|
Severe acute respiratory syndrome coronavirus 2
|
12.91
%
|
|
SARS-CoV-2 3CL-Pro protease inhibition percentage at 20µM by FRET kind of response from peptide substrate
|
Severe acute respiratory syndrome coronavirus 2
|
10.6
%
|
|
|
Antiviral activity determined as inhibition of SARS-CoV-2 induced cytotoxicity of VERO-6 cells at 10 uM after 48 hours exposure to 0.01 MOI SARS CoV-2 virus by high content imaging
|
Chlorocebus sabaeus
|
-0.32
%
|
|
Antiviral activity determined as inhibition of SARS-CoV-2 induced cytotoxicity of VERO-6 cells at 10 uM after 48 hours exposure to 0.01 MOI SARS CoV-2 virus by high content imaging
|
Chlorocebus sabaeus
|
-0.33
%
|
|
Antiviral activity determined as inhibition of SARS-CoV-2 induced cytotoxicity of VERO-6 cells at 10 uM after 48 hours exposure to 0.01 MOI SARS CoV-2 virus by high content imaging
|
Chlorocebus sabaeus
|
-0.33
%
|
|
Antiviral activity determined as inhibition of SARS-CoV-2 induced cytotoxicity of VERO-6 cells at 10 uM after 48 hours exposure to 0.01 MOI SARS CoV-2 virus by high content imaging
|
Chlorocebus sabaeus
|
-0.32
%
|
|