| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| ATC | A11CC03 |
| UNII | URQ2517572 |
| EPA CompTox | DTXSID0022569 |
Structure
| InChI Key | OFHCOWSQAMBJIW-AVJTYSNKSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C27H44O2 |
| Molecular Weight | 400.65 |
| AlogP | 6.59 |
| Hydrogen Bond Acceptor | 2.0 |
| Hydrogen Bond Donor | 2.0 |
| Number of Rotational Bond | 6.0 |
| Polar Surface Area | 40.46 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 0.0 |
| Heavy Atoms | 29.0 |
Pharmacology
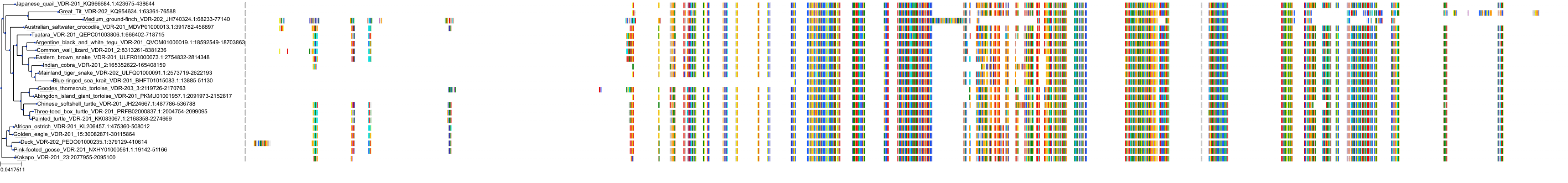
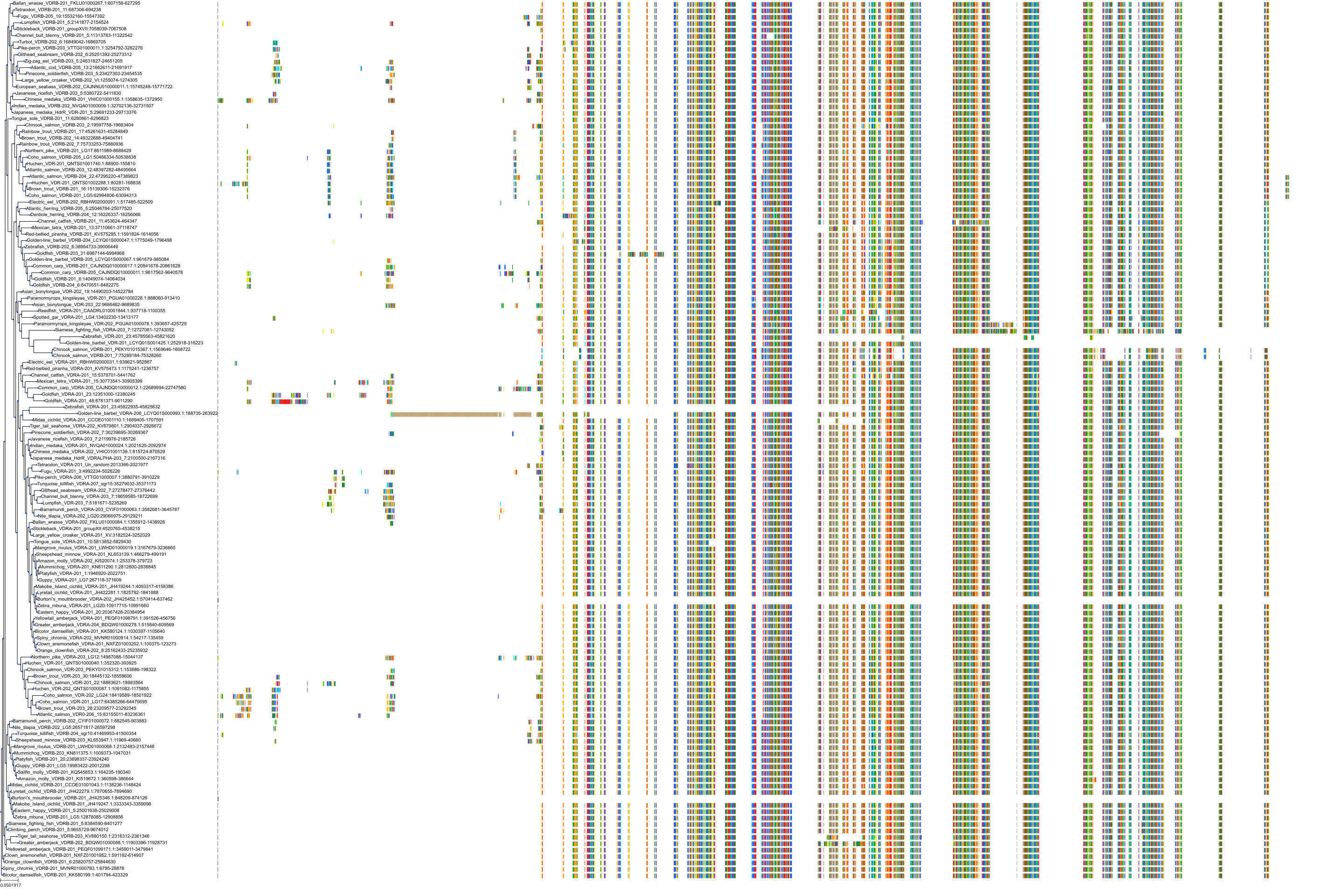
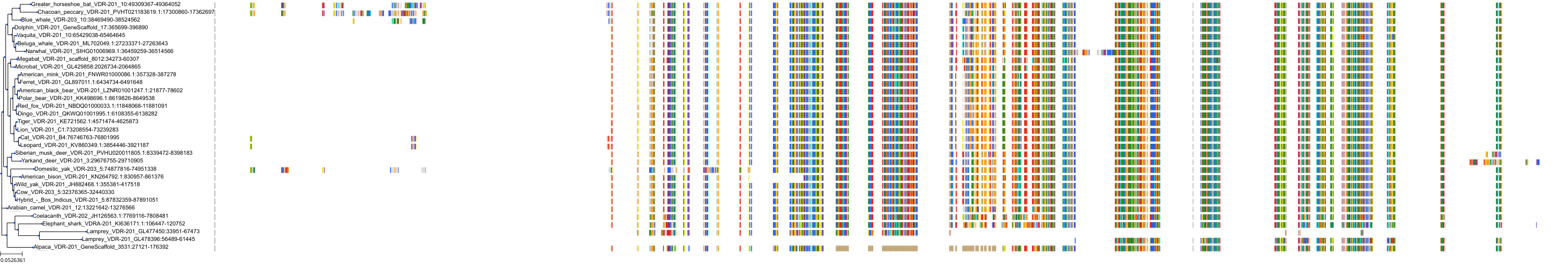
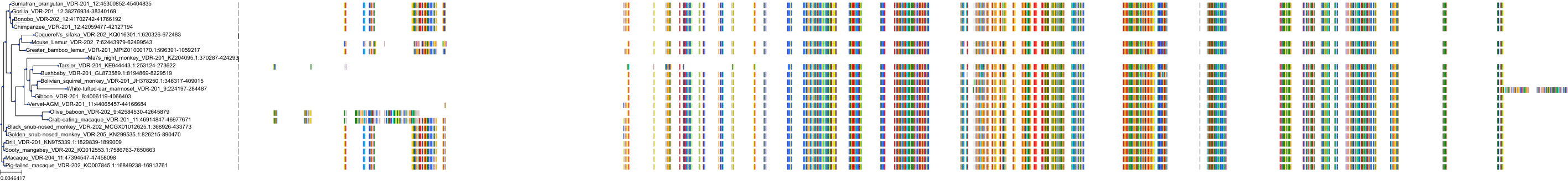
Target Conservation
|
Protein: Vitamin D receptor Description: Vitamin D3 receptor Organism : Homo sapiens P11473 ENSG00000111424 |
||||
Related Entries
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 31186 |
| ChEMBL | CHEMBL1601669 |
| DrugBank | DB01436 |
| DrugCentral | 130 |
| FDA SRS | URQ2517572 |
| Human Metabolome Database | HMDB0015504 |
| PDB | M9B |
| PharmGKB | PA164746469 |
| PubChem | 5282181 |
| SureChEMBL | SCHEMBL3063 |
| ZINC | ZINC000012484965 |