| Trade Names | |
| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| ATC | B01AC21 |
| UNII | RUM6K67ESG |
Structure
| InChI Key | PAJMKGZZBBTTOY-ZFORQUDYSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C23H34O5 |
| Molecular Weight | 390.52 |
| AlogP | 3.58 |
| Hydrogen Bond Acceptor | 4.0 |
| Hydrogen Bond Donor | 3.0 |
| Number of Rotational Bond | 10.0 |
| Polar Surface Area | 86.99 |
| Molecular species | ACID |
| Aromatic Rings | 1.0 |
| Heavy Atoms | 28.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Prostanoid IP receptor agonist | AGONIST | PubMed |
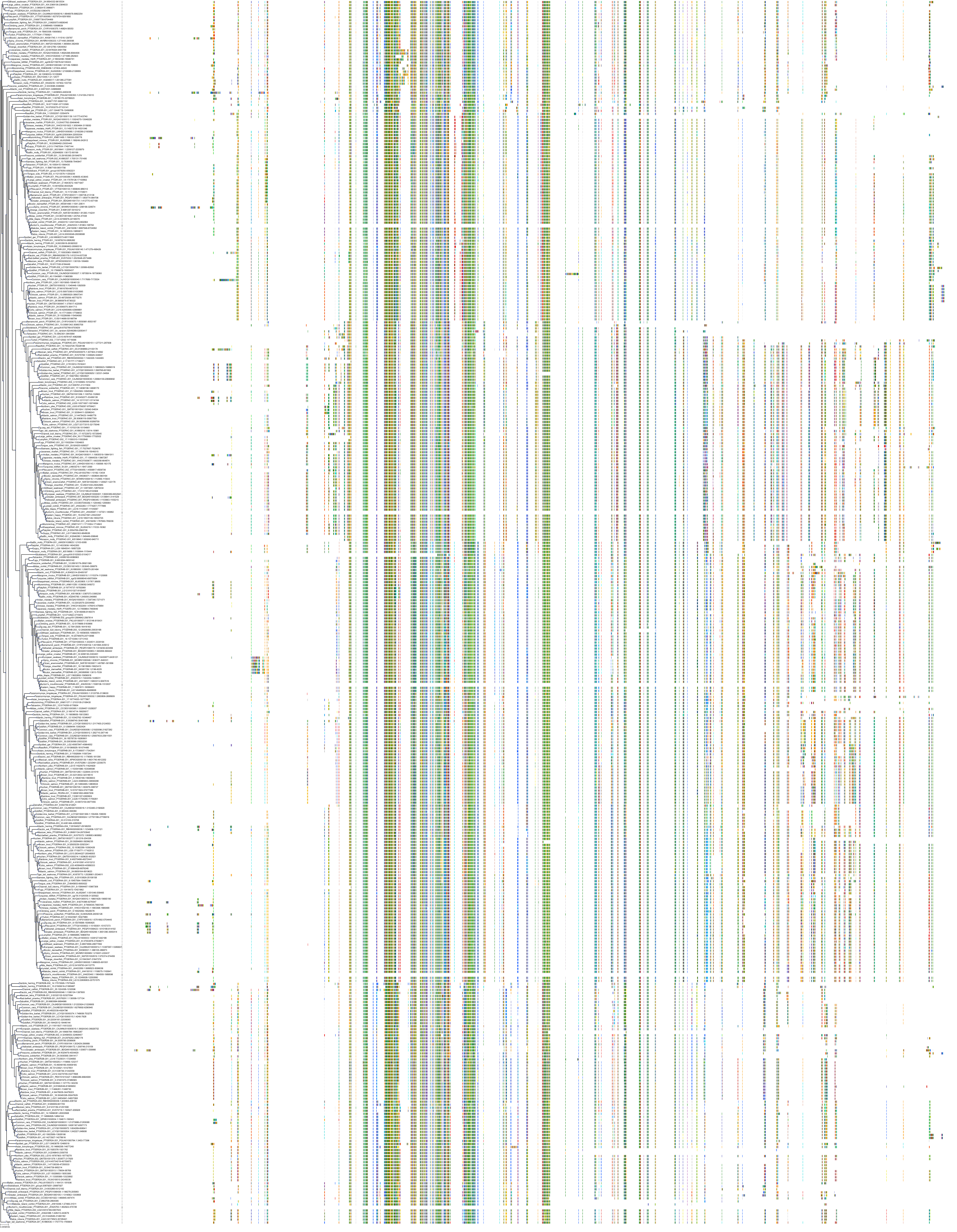
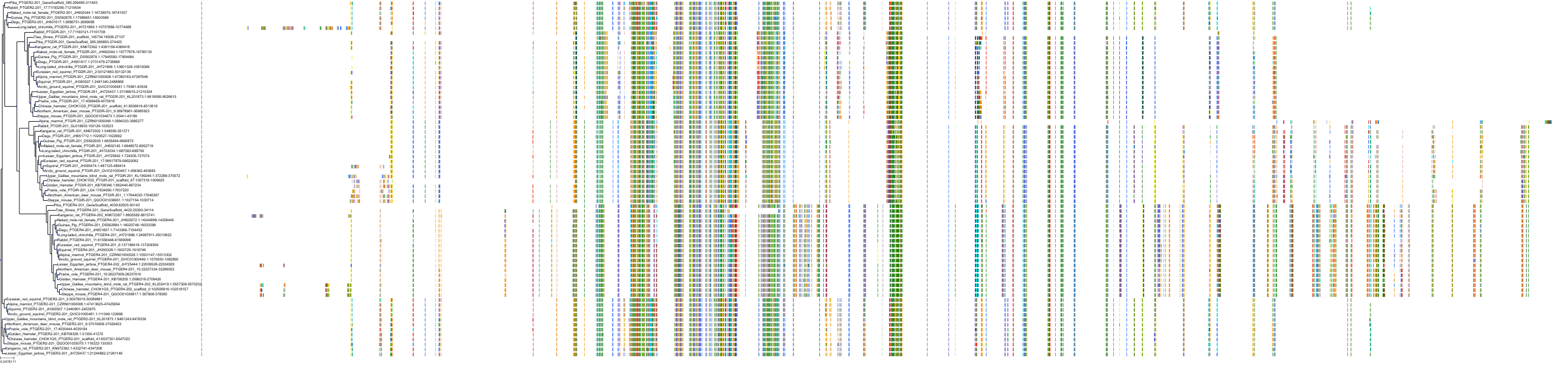
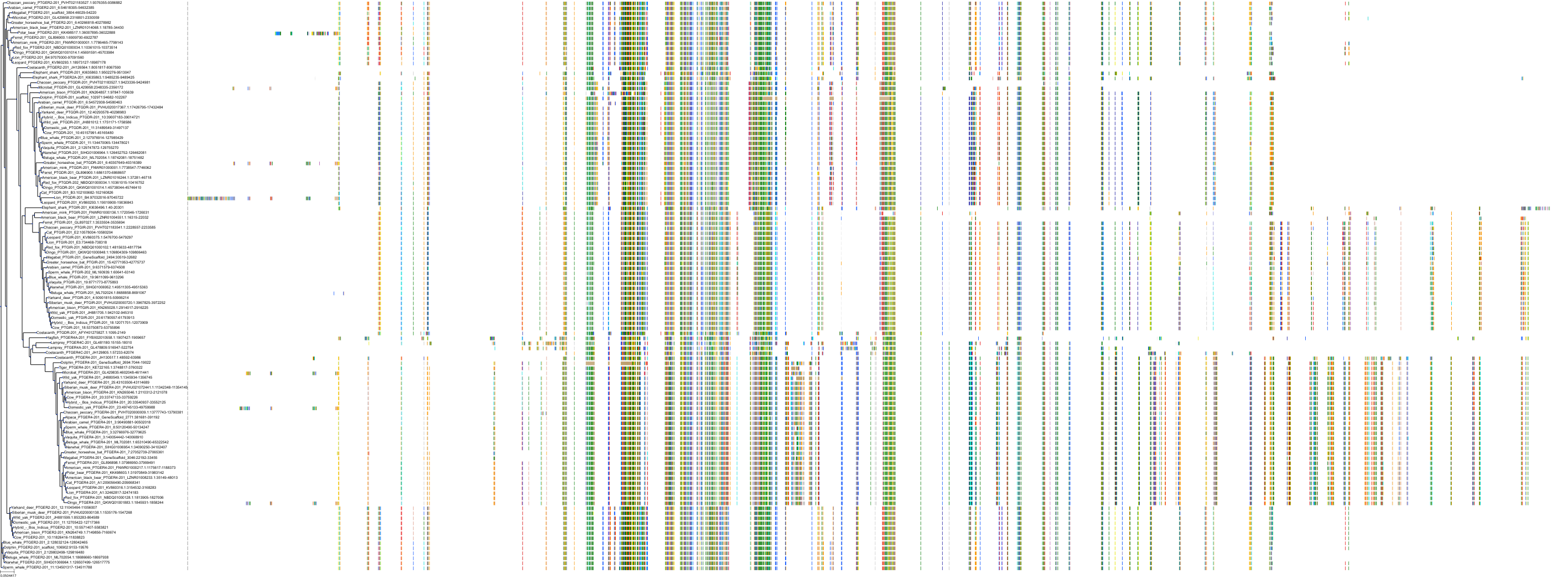
Target Conservation
|
Protein: Prostanoid IP receptor Description: Prostacyclin receptor Organism : Homo sapiens P43119 ENSG00000160013 |
||||
Related Entries
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 50861 |
| ChEMBL | CHEMBL1237119 |
| DrugBank | DB00374 |
| DrugCentral | 2720 |
| FDA SRS | RUM6K67ESG |
| Human Metabolome Database | HMDB0014518 |
| Guide to Pharmacology | 5820 |
| PubChem | 6918140 |
| SureChEMBL | SCHEMBL4349618 |
| ZINC | ZINC000003800475 |