| Trade Names | |
| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| ATC | B01AC24 |
| UNII | GLH0314RVC |
Structure
| InChI Key | OEKWJQXRCDYSHL-FNOIDJSQSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C23H28F2N6O4S |
| Molecular Weight | 522.58 |
| AlogP | 2.01 |
| Hydrogen Bond Acceptor | 11.0 |
| Hydrogen Bond Donor | 4.0 |
| Number of Rotational Bond | 10.0 |
| Polar Surface Area | 138.44 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 3.0 |
| Heavy Atoms | 36.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Purinergic receptor P2Y12 negative allosteric modulator | NEGATIVE ALLOSTERIC MODULATOR | FDA |
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Membrane receptor
Family A G protein-coupled receptor
Small molecule receptor (family A GPCR)
Nucleotide-like receptor (family A GPCR)
Purine receptor
|
- | 5.012-500 | - | 1.995-14 | - |
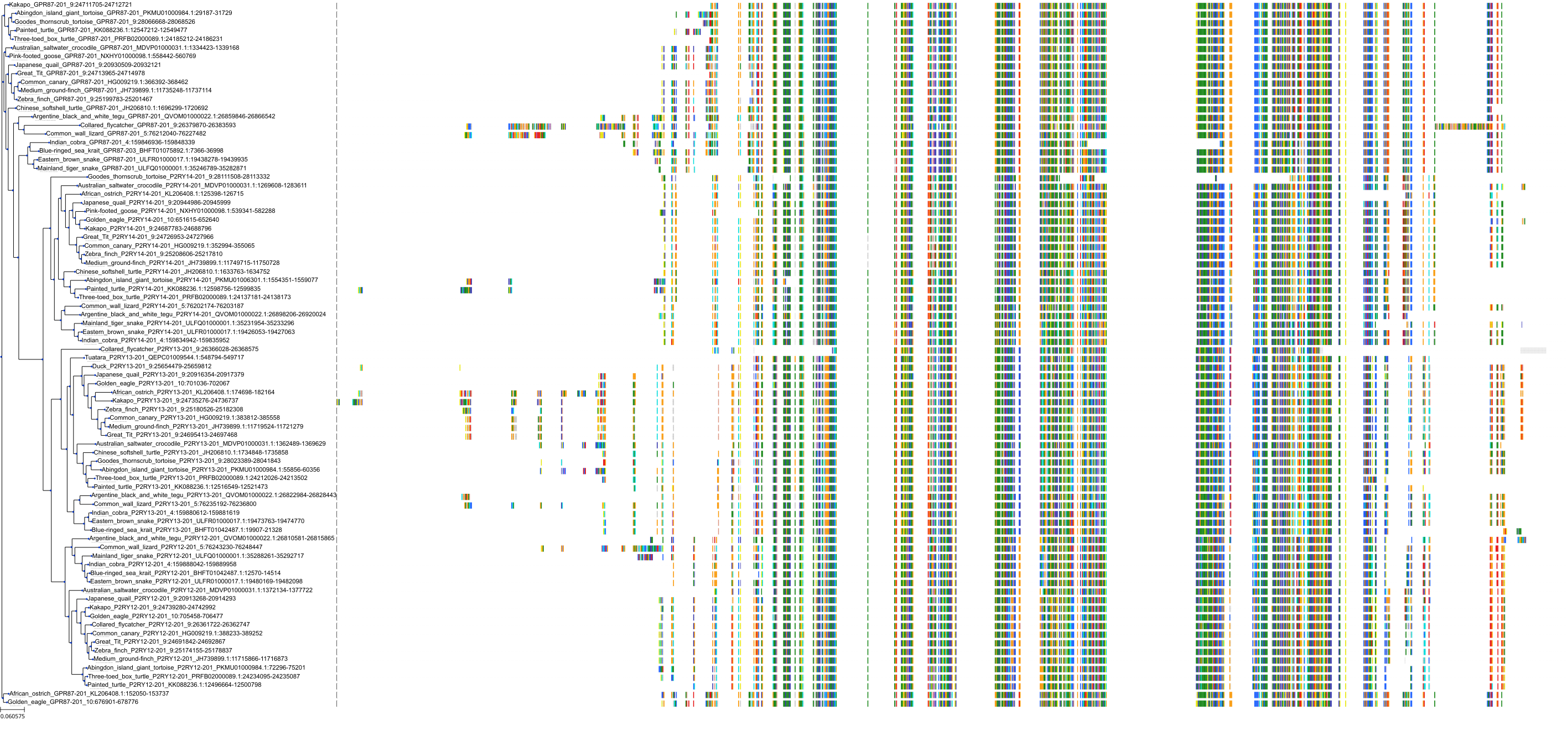
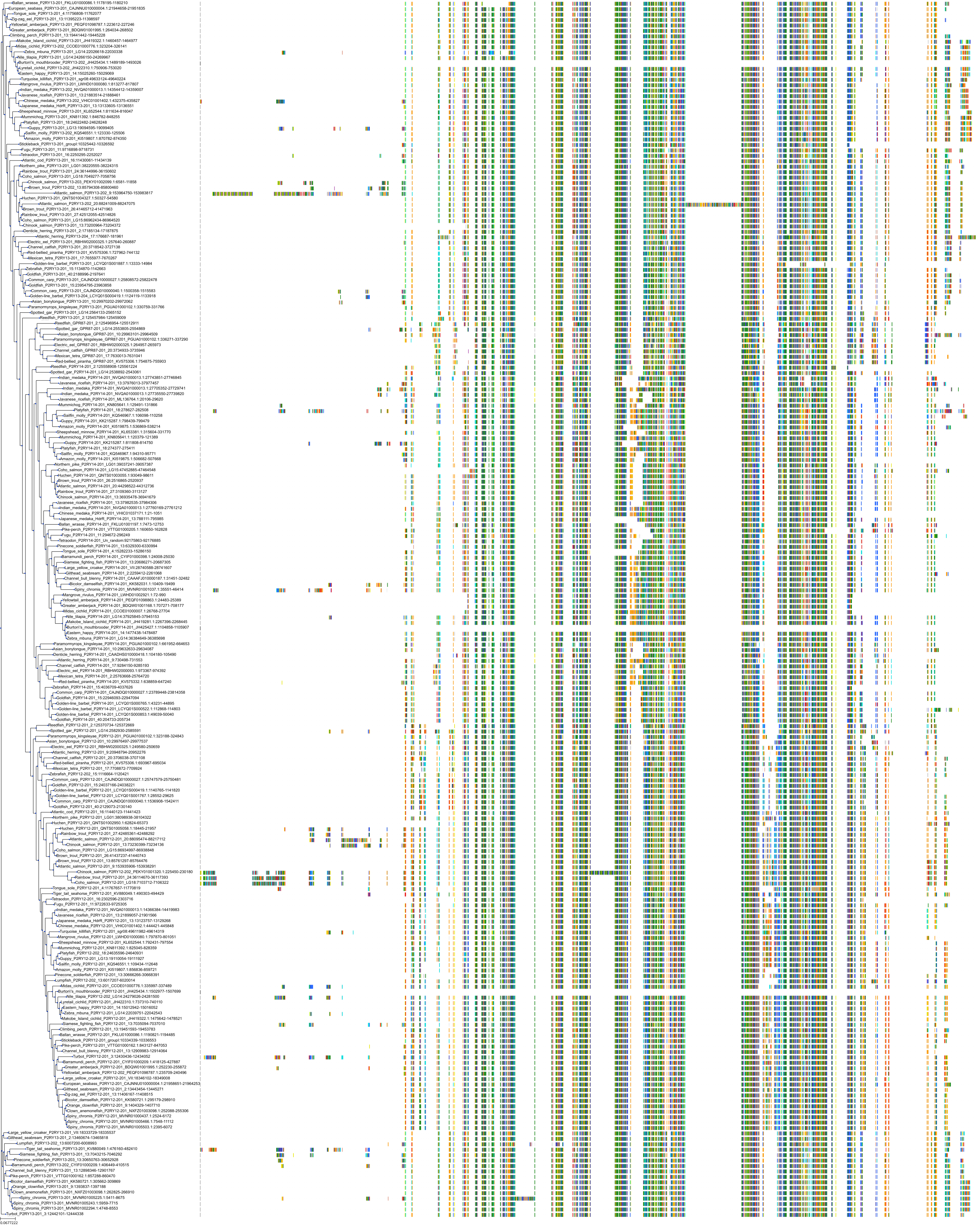
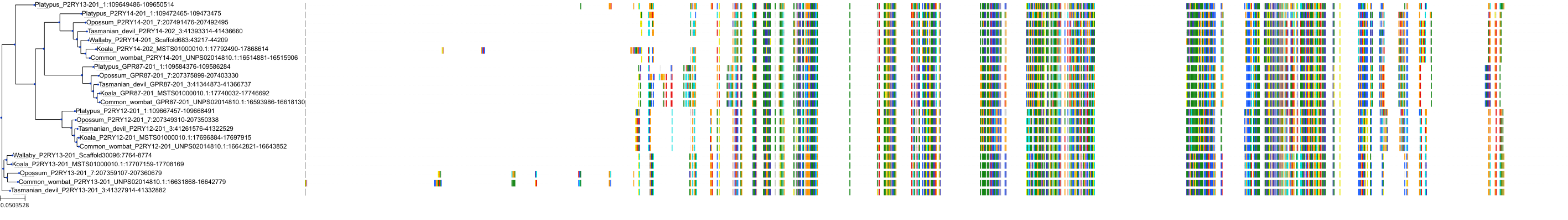
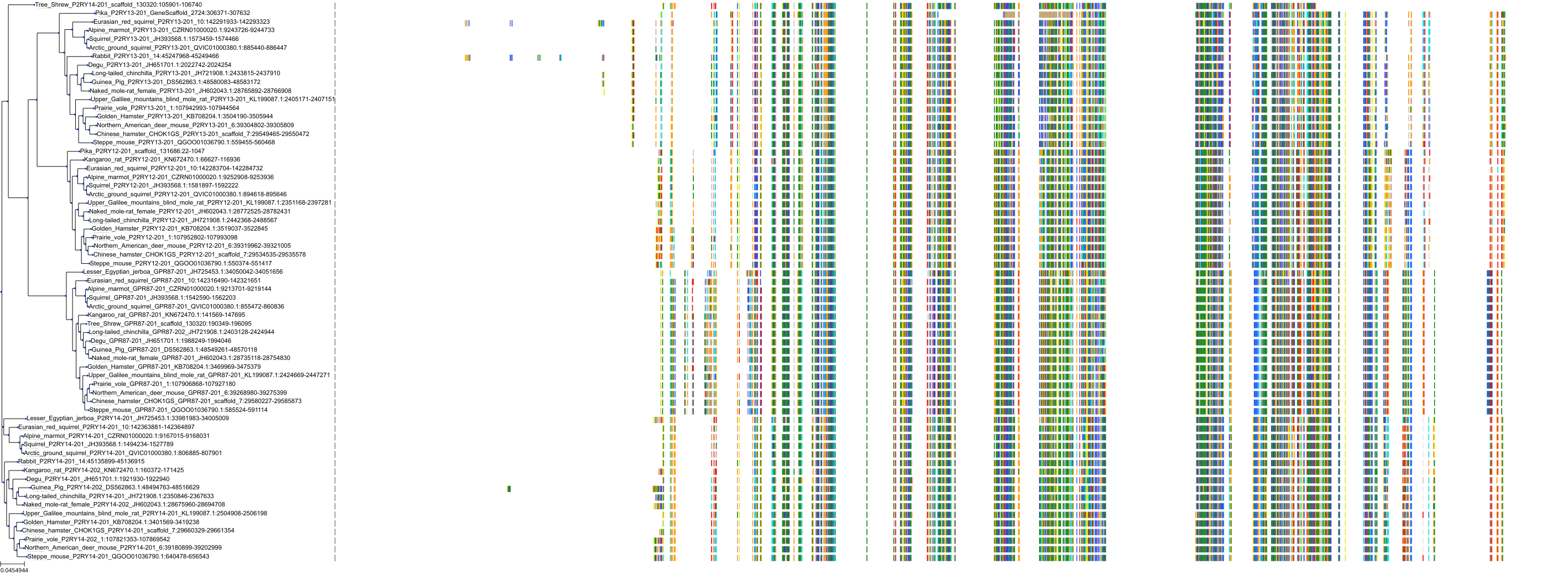
Target Conservation
|
Protein: Purinergic receptor P2Y12 Description: P2Y purinoceptor 12 Organism : Homo sapiens Q9H244 ENSG00000169313 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 68558 |
| ChEMBL | CHEMBL398435 |
| DrugBank | DB08816 |
| DrugCentral | 4184 |
| FDA SRS | GLH0314RVC |
| Human Metabolome Database | HMDB0015702 |
| Guide to Pharmacology | 1765 |
| KEGG | D09017 |
| PDB | TIQ |
| PharmGKB | PA165374673 |
| PubChem | 9871419 |
| SureChEMBL | SCHEMBL1979652 |
| ZINC | ZINC000028957444 |
 Homo sapiens
Homo sapiens
 Rattus norvegicus
Rattus norvegicus