| Trade Names | |
| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| ATC | G04CA04 |
| UNII | CUZ39LUY82 |
| EPA CompTox | DTXSID40167045 |
Structure
| InChI Key | PNCPYILNMDWPEY-QGZVFWFLSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C25H32F3N3O4 |
| Molecular Weight | 495.54 |
| AlogP | 3.07 |
| Hydrogen Bond Acceptor | 6.0 |
| Hydrogen Bond Donor | 3.0 |
| Number of Rotational Bond | 13.0 |
| Polar Surface Area | 97.05 |
| Molecular species | BASE |
| Aromatic Rings | 2.0 |
| Heavy Atoms | 35.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Alpha-1a adrenergic receptor antagonist | ANTAGONIST | DailyMed |
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Membrane receptor
Family A G protein-coupled receptor
Small molecule receptor (family A GPCR)
Monoamine receptor
Adrenergic receptor
|
- | 0.8-541.4 | - | 0.036-19.95 | 100 |
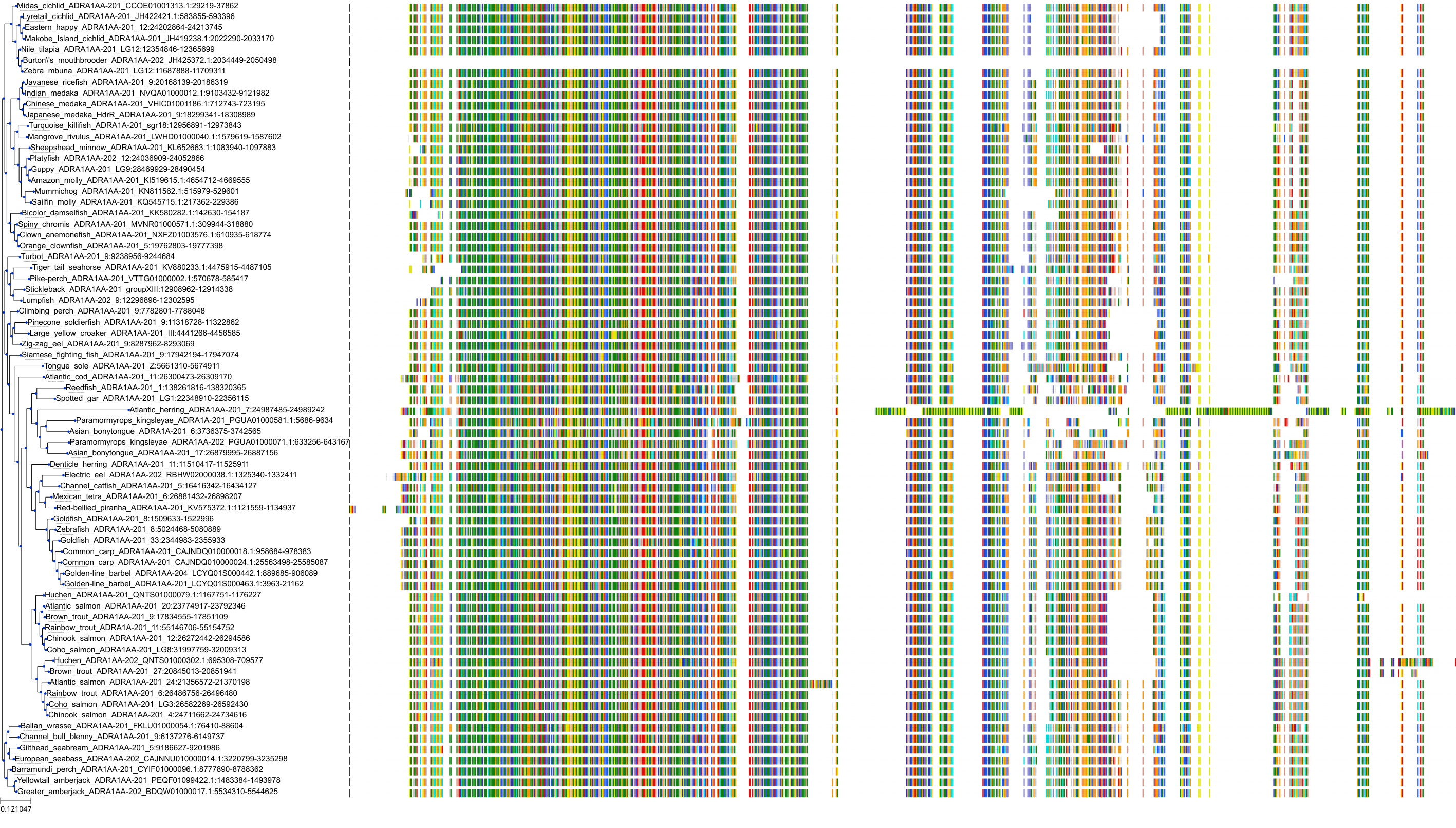
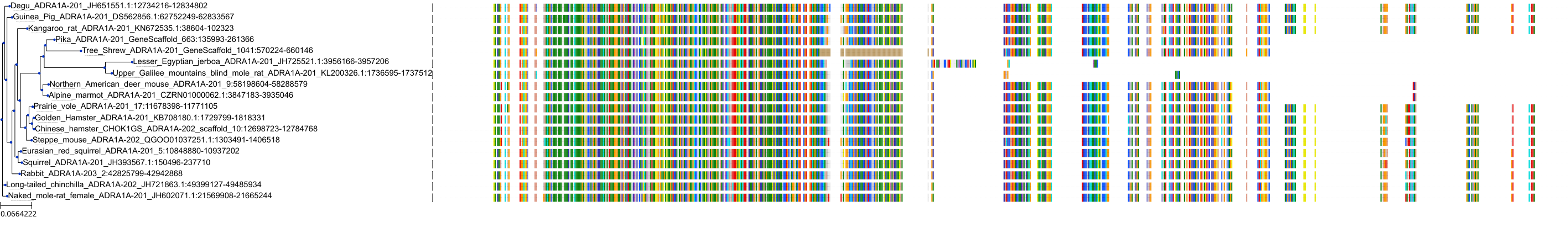
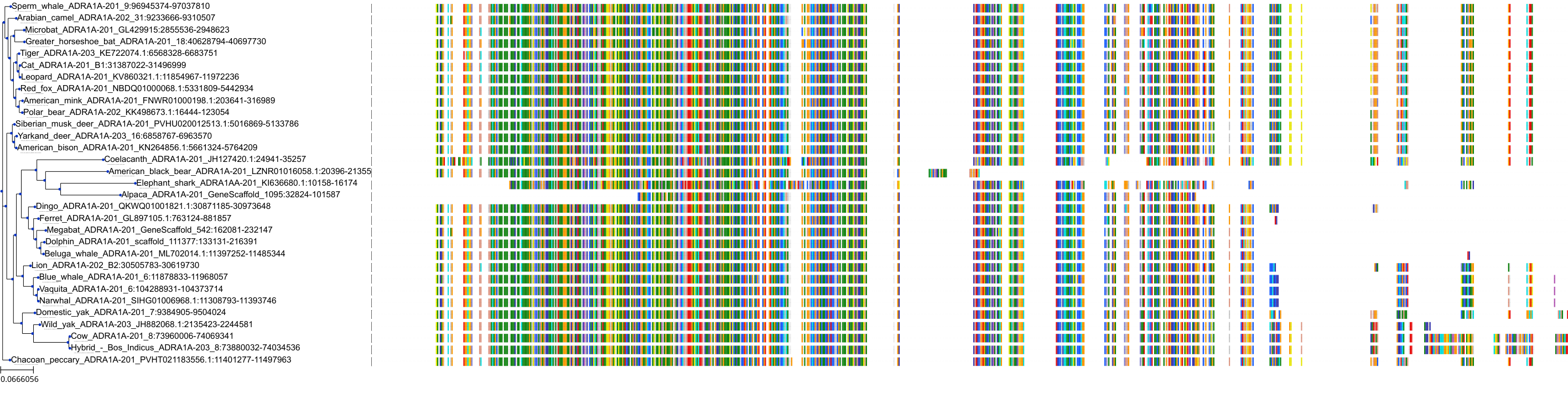
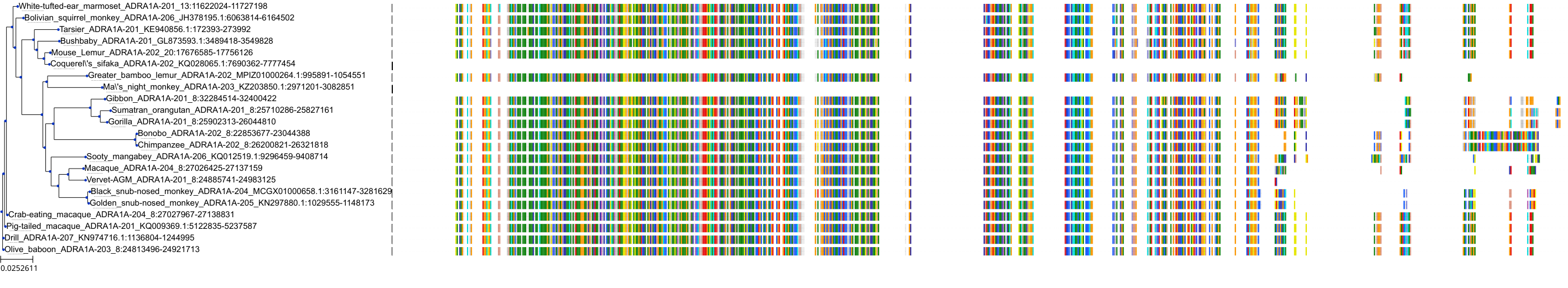
Target Conservation
|
Protein: Alpha-1a adrenergic receptor Description: Alpha-1A adrenergic receptor Organism : Homo sapiens P35348 ENSG00000120907 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 135929 |
| ChEMBL | CHEMBL24778 |
| DrugBank | DB06207 |
| DrugCentral | 4151 |
| FDA SRS | CUZ39LUY82 |
| Guide to Pharmacology | 493 |
| KEGG | D01965 |
| PubChem | 5312125 |
| SureChEMBL | SCHEMBL136973 |
| ZINC | ZINC000003806063 |
 Homo sapiens
Homo sapiens
 Rattus norvegicus
Rattus norvegicus