| Trade Names | |
| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| ATC | N05CH02 |
| UNII | 901AS54I69 |
Structure
| InChI Key | YLXDSYKOBKBWJQ-LBPRGKRZSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C16H21NO2 |
| Molecular Weight | 259.35 |
| AlogP | 2.57 |
| Hydrogen Bond Acceptor | 2.0 |
| Hydrogen Bond Donor | 1.0 |
| Number of Rotational Bond | 4.0 |
| Polar Surface Area | 38.33 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 1.0 |
| Heavy Atoms | 19.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Melatonin receptor agonist | AGONIST | FDA |
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Membrane receptor
Family A G protein-coupled receptor
Small molecule receptor (family A GPCR)
Monoamine-derivative receptor (family A GPCR)
Melatonin receptor
|
- | - | - | 0.0138-0.112 | - | |
|
Transporter
Electrochemical transporter
SLC superfamily of solute carriers
SLC21/SLCO family of organic anion transporting polypeptides
|
- | - | - | - | 113.38-127.58 |
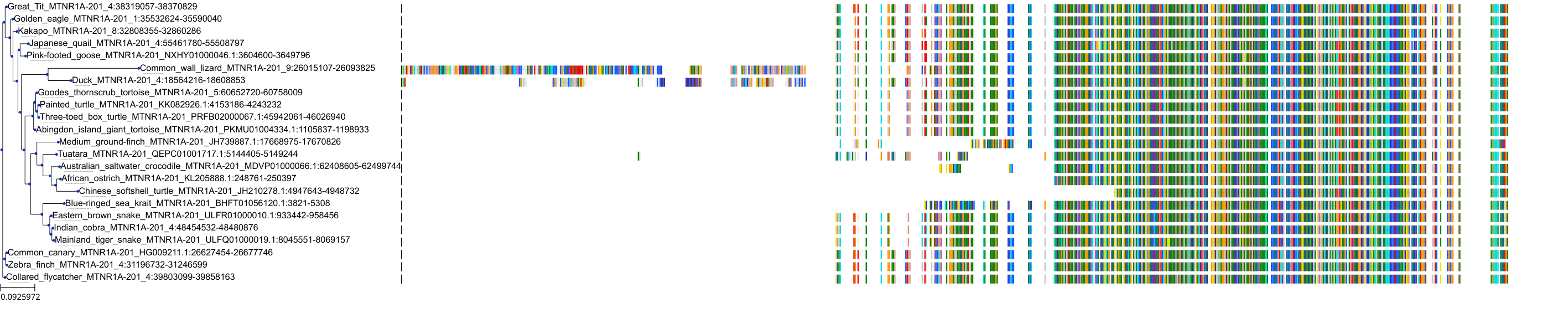
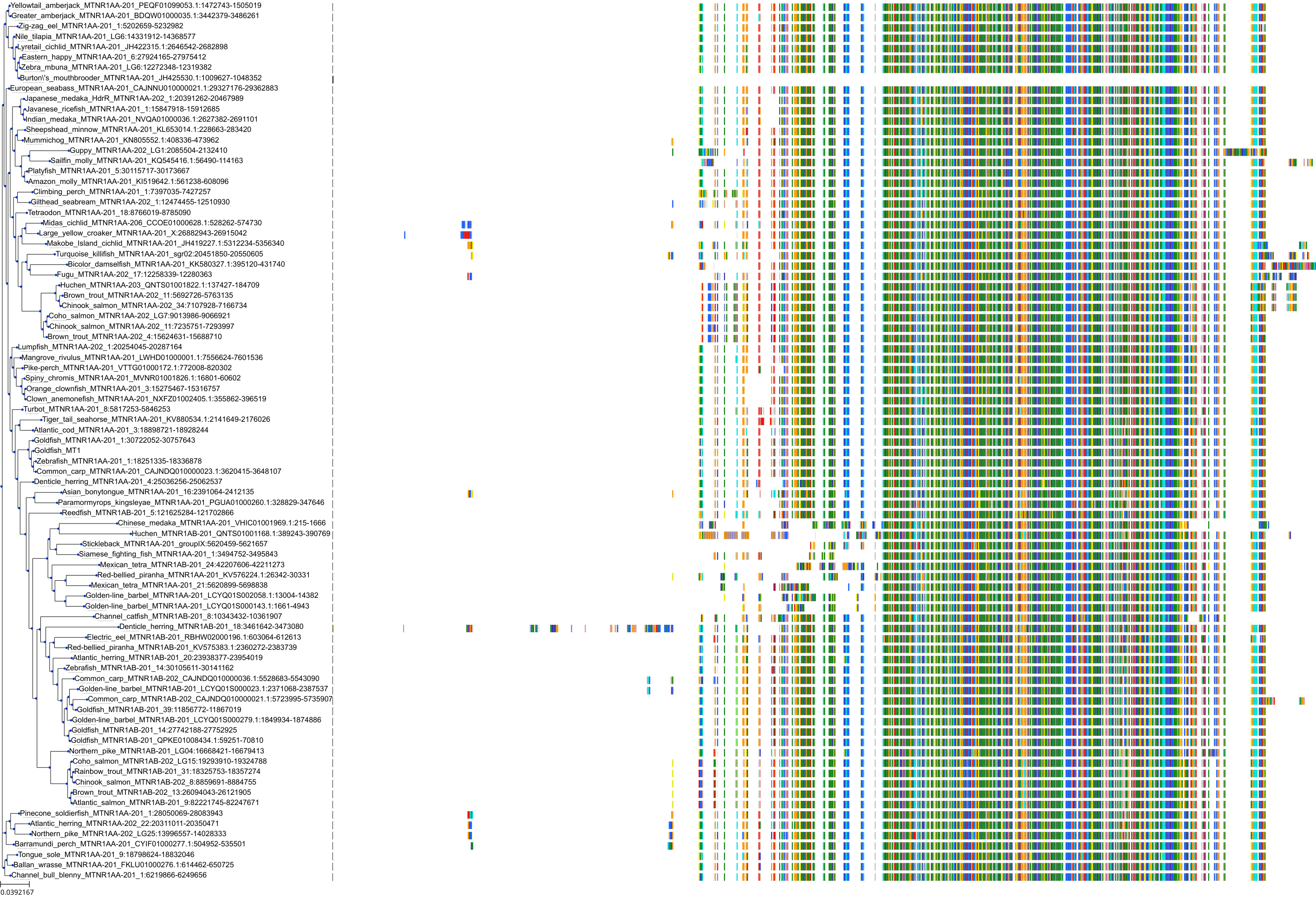
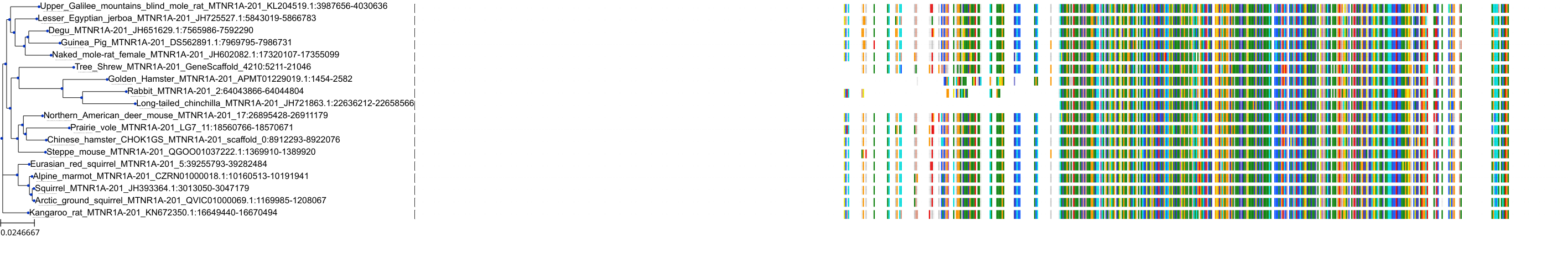
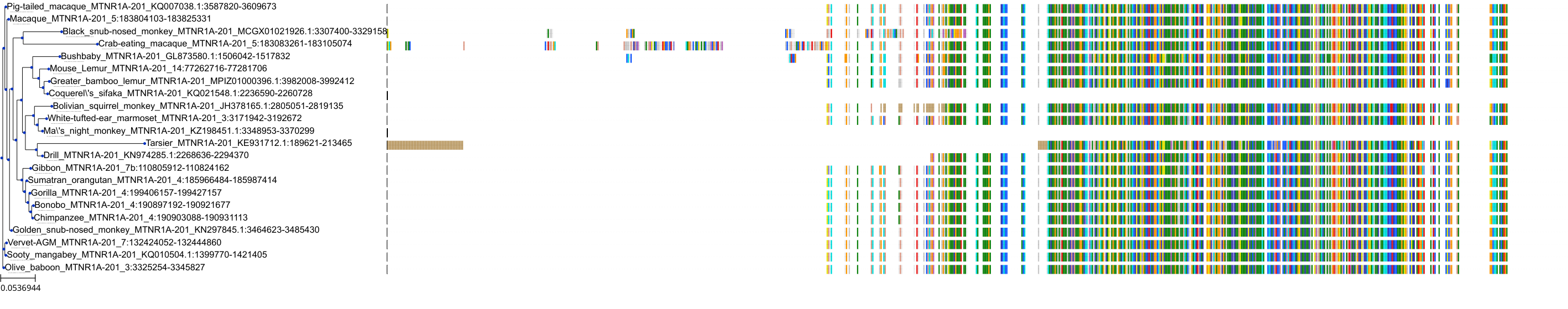
Target Conservation
|
Protein: Melatonin receptor Description: Melatonin receptor type 1A Organism : Homo sapiens P48039 ENSG00000168412 |
||||
|
Protein: Melatonin receptor Description: Melatonin receptor type 1B Organism : Homo sapiens P49286 ENSG00000134640 |
||||
Related Entries
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 109549 |
| ChEMBL | CHEMBL1218 |
| DrugBank | DB00980 |
| DrugCentral | 2355 |
| FDA SRS | 901AS54I69 |
| Human Metabolome Database | HMDB0015115 |
| Guide to Pharmacology | 1356 |
| KEGG | D02689 |
| PDB | JEV |
| PharmGKB | PA164744896 |
| PubChem | 208902 |
| SureChEMBL | SCHEMBL29237 |
| ZINC | ZINC000003960338 |
 Cricetulus griseus
Cricetulus griseus
 Homo sapiens
Homo sapiens
 Rattus norvegicus
Rattus norvegicus