| Trade Names | |
| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| ATC | N07BA01 |
| UNII | 6M3C89ZY6R |
| EPA CompTox | DTXSID1020930 |
Structure
| InChI Key | SNICXCGAKADSCV-JTQLQIEISA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C10H14N2 |
| Molecular Weight | 162.24 |
| AlogP | 1.85 |
| Hydrogen Bond Acceptor | 2.0 |
| Number of Rotational Bond | 1.0 |
| Polar Surface Area | 16.13 |
| Molecular species | BASE |
| Aromatic Rings | 1.0 |
| Heavy Atoms | 12.0 |
Metabolites Network
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Neuronal acetylcholine receptor; alpha4/beta2 agonist | AGONIST | PubMed PubMed PubMed PubMed PubMed PubMed PubMed PubMed PubMed PubMed PubMed |
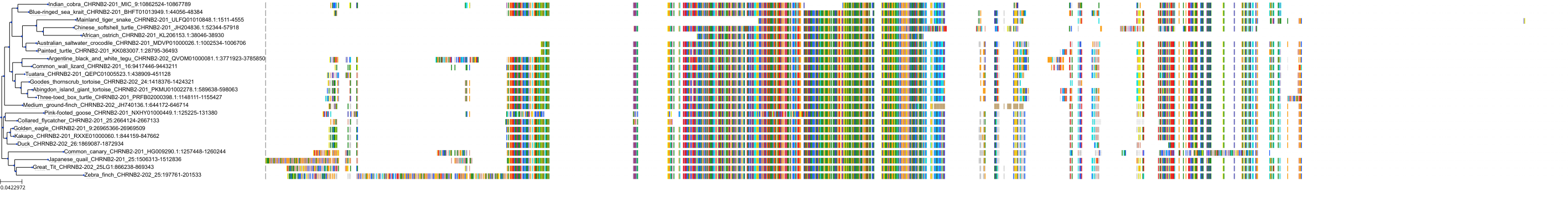
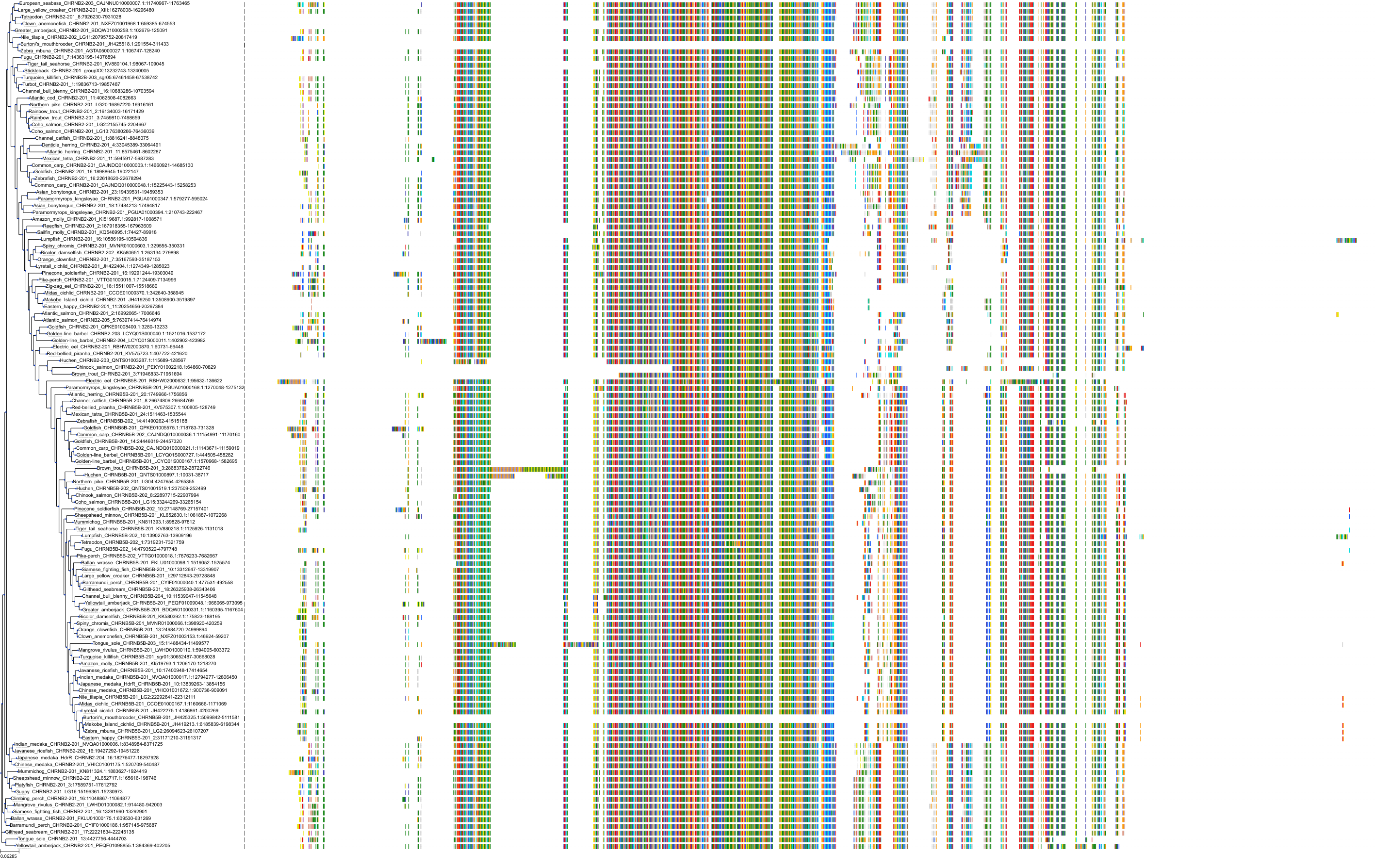
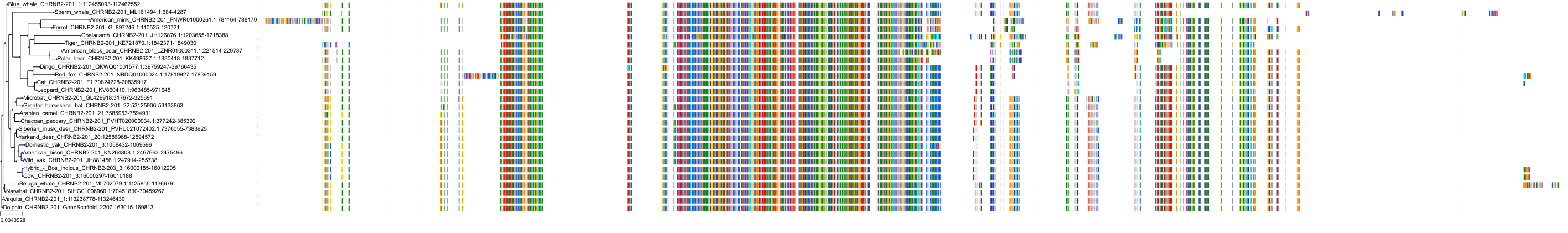
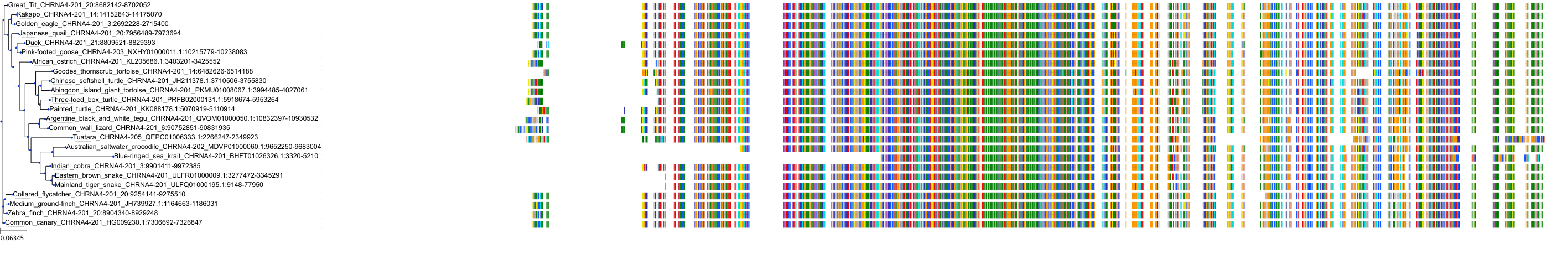
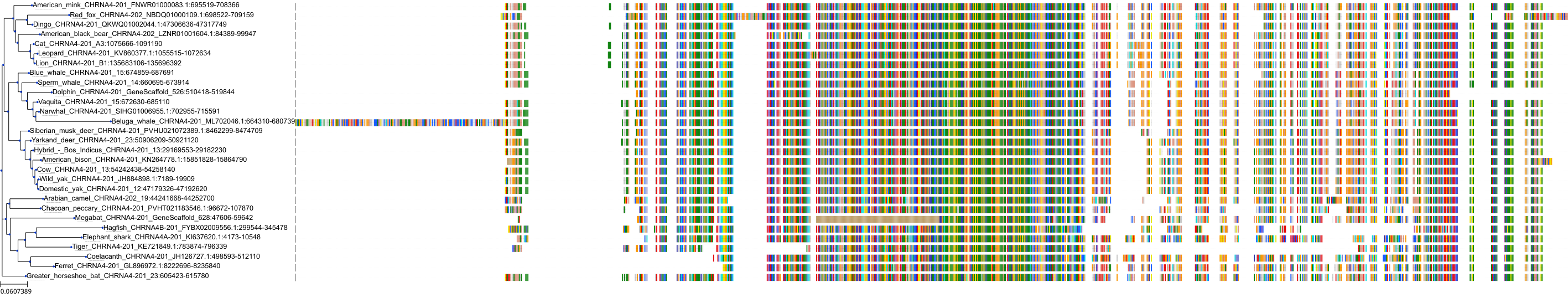
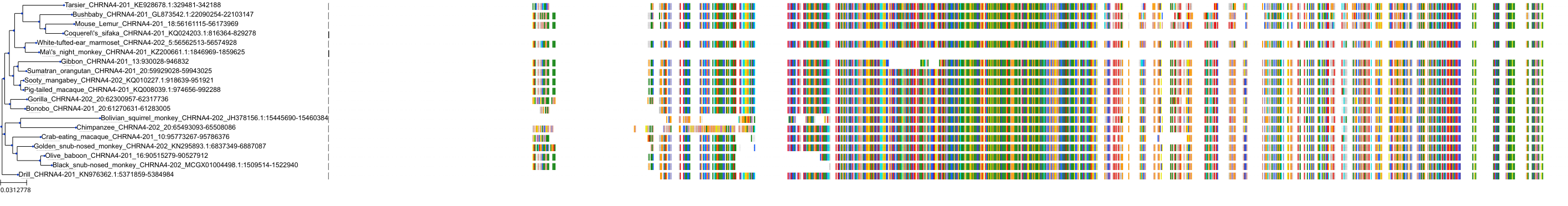
Target Conservation
|
Protein: Neuronal acetylcholine receptor; alpha4/beta2 Description: Neuronal acetylcholine receptor subunit beta-2 Organism : Homo sapiens P17787 ENSG00000160716 |
||||
|
Protein: Neuronal acetylcholine receptor; alpha4/beta2 Description: Neuronal acetylcholine receptor subunit alpha-4 Organism : Homo sapiens P43681 ENSG00000101204 |
||||
Related Entries
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 17688 |
| ChEMBL | CHEMBL3 |
| DrugBank | DB00184 |
| DrugCentral | 1920 |
| FDA SRS | 6M3C89ZY6R |
| Human Metabolome Database | HMDB0001934 |
| Guide to Pharmacology | 2585 |
| KEGG | C00745 |
| PDB | NCT |
| PubChem | 89594 |
| SureChEMBL | SCHEMBL20192 |
| ZINC | ZINC000000391812 |
 Aplysia
Aplysia
 Aplysia californica
Aplysia californica
 Bos taurus
Bos taurus
 Electrophorus electricus
Electrophorus electricus
 Equus caballus
Equus caballus
 Gallus gallus
Gallus gallus
 Homo sapiens
Homo sapiens
 Lymnaea stagnalis
Lymnaea stagnalis
 Mus musculus
Mus musculus
 Myzus persicae
Myzus persicae
 Rattus norvegicus
Rattus norvegicus
 Sus scrofa
Sus scrofa
 Torpedo
Torpedo