| Trade Names | |
| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| ATC | D10AD04 D10BA01 |
| UNII | EH28UP18IF |
| EPA CompTox | DTXSID4023177 |
Structure
| InChI Key | SHGAZHPCJJPHSC-XFYACQKRSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C20H28O2 |
| Molecular Weight | 300.44 |
| AlogP | 5.6 |
| Hydrogen Bond Acceptor | 1.0 |
| Hydrogen Bond Donor | 1.0 |
| Number of Rotational Bond | 5.0 |
| Polar Surface Area | 37.3 |
| Molecular species | ACID |
| Aromatic Rings | 0.0 |
| Heavy Atoms | 22.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Retinoic acid receptor agonist | AGONIST | PubMed |
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Enzyme
Hydrolase
|
- | - | - | - | 0.91-11.5 | |
|
Enzyme
Lyase
|
- | - | - | - | 72-96 | |
|
Other cytosolic protein
|
- | 900-900 | - | - | - | |
|
Transporter
Electrochemical transporter
SLC superfamily of solute carriers
SLC21/SLCO family of organic anion transporting polypeptides
|
- | - | - | - | 32.24-51.47 |
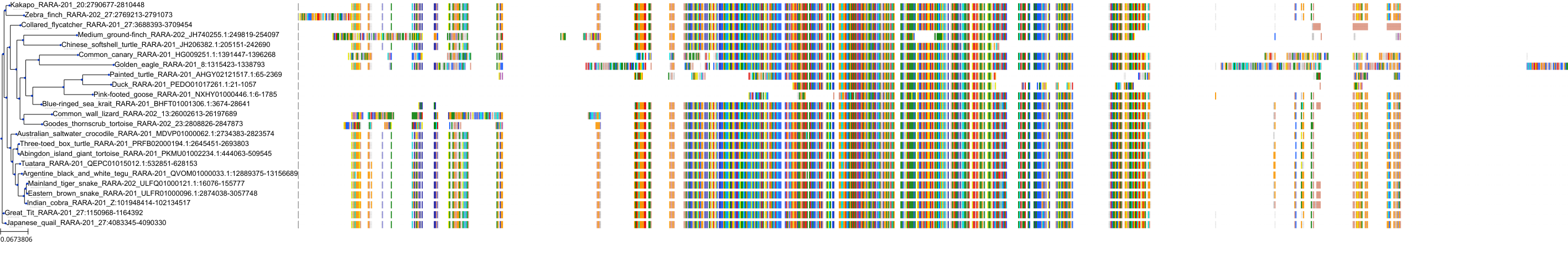
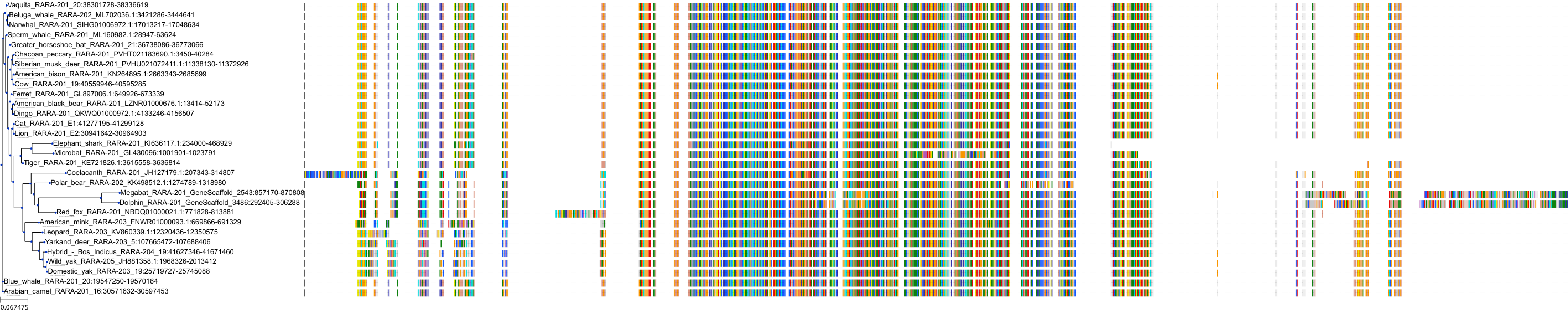
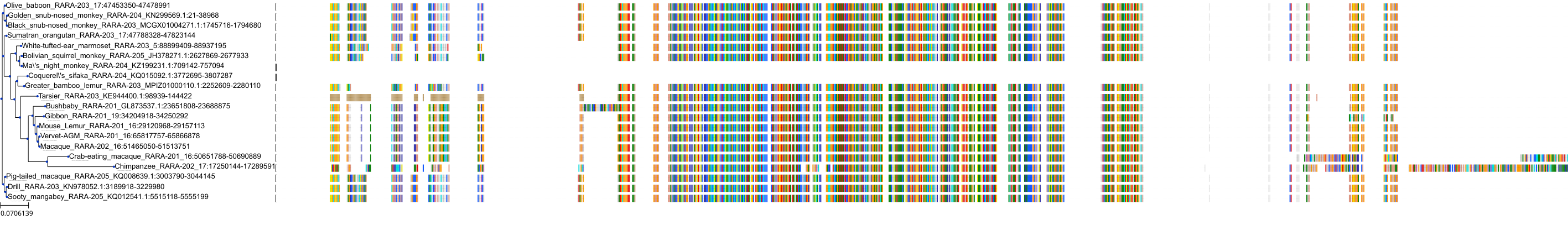
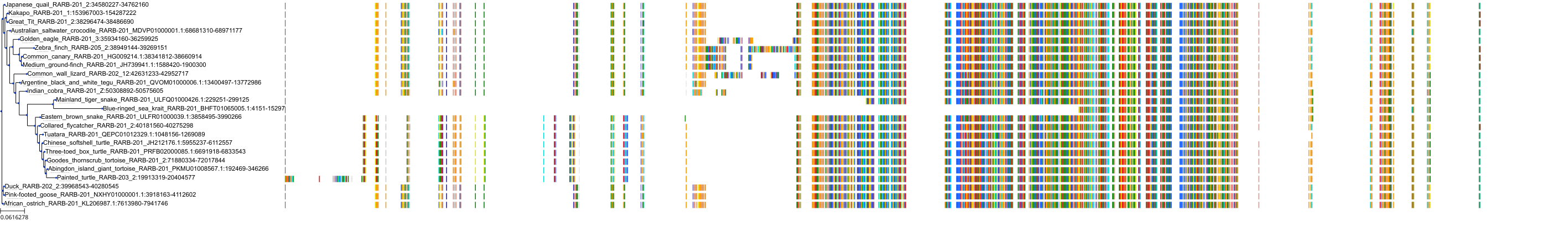
Target Conservation
|
Protein: Retinoic acid receptor Description: Retinoic acid receptor alpha Organism : Homo sapiens P10276 ENSG00000131759 |
||||
|
Protein: Retinoic acid receptor Description: Retinoic acid receptor beta Organism : Homo sapiens P10826 ENSG00000077092 |
||||
|
Protein: Retinoic acid receptor Description: Retinoic acid receptor gamma Organism : Homo sapiens P13631 ENSG00000172819 |
||||
Related Entries
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 6067 |
| ChEMBL | CHEMBL547 |
| DrugBank | DB00982 |
| DrugCentral | 1508 |
| FDA SRS | EH28UP18IF |
| Human Metabolome Database | HMDB0006219 |
| Guide to Pharmacology | 7600 |
| KEGG | C07058 |
| PDB | REA |
| PubChem | 5282379 |
| SureChEMBL | SCHEMBL38299 |
| ZINC | ZINC000003792789 |
 Cricetulus griseus
Cricetulus griseus
 Electrophorus electricus
Electrophorus electricus
 Equus caballus
Equus caballus
 Gallus gallus
Gallus gallus
 Homo sapiens
Homo sapiens
 Mus musculus
Mus musculus
 Rattus norvegicus
Rattus norvegicus