| Trade Names | |
| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| ATC | R06AX27 |
| UNII | FVF865388R |
| EPA CompTox | DTXSID1044196 |
Structure
| InChI Key | JAUOIFJMECXRGI-UHFFFAOYSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C19H19ClN2 |
| Molecular Weight | 310.83 |
| AlogP | 4.02 |
| Hydrogen Bond Acceptor | 2.0 |
| Hydrogen Bond Donor | 1.0 |
| Polar Surface Area | 24.92 |
| Molecular species | BASE |
| Aromatic Rings | 2.0 |
| Heavy Atoms | 22.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Histamine H1 receptor antagonist | ANTAGONIST | DailyMed |
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Membrane receptor
Family A G protein-coupled receptor
Small molecule receptor (family A GPCR)
Monoamine receptor
Histamine receptor
|
- | 1.84 | 24 | 0.97-1.585 | 47.42-80.44 |
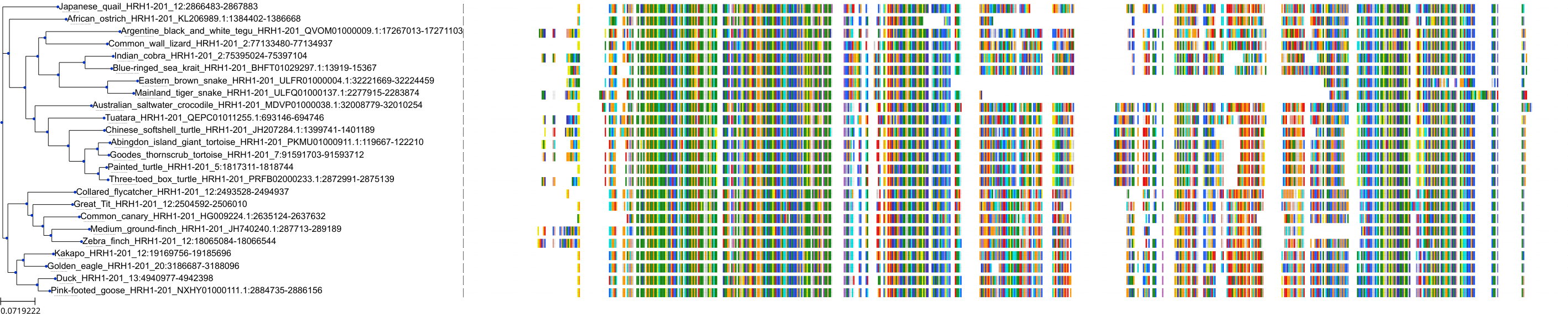
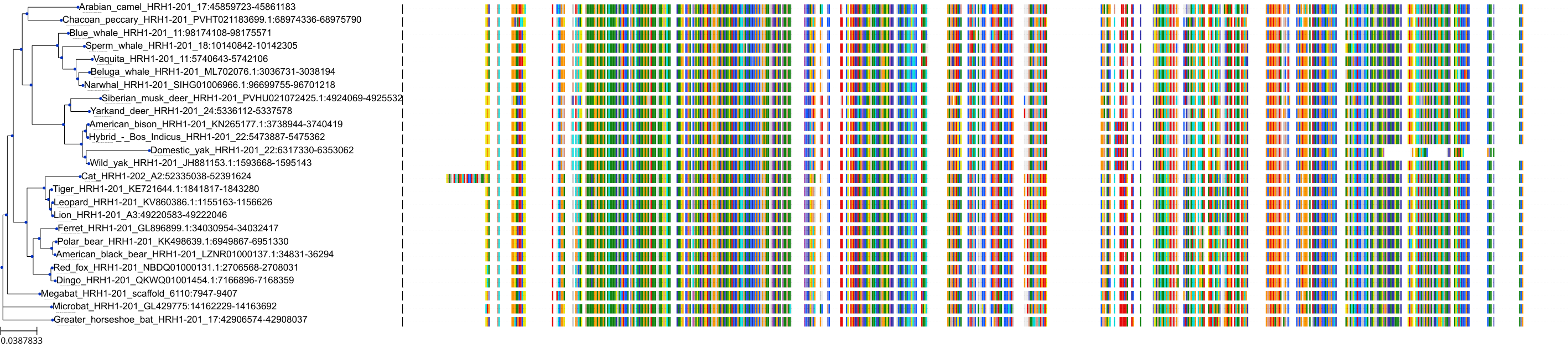
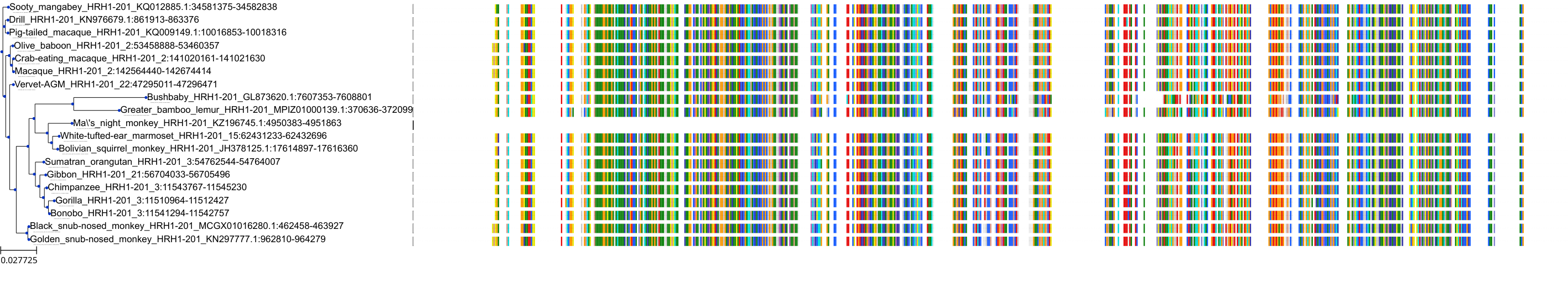
Target Conservation
|
Protein: Histamine H1 receptor Description: Histamine H1 receptor Organism : Homo sapiens P35367 ENSG00000196639 |
||||
Related Entries
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 291342 |
| ChEMBL | CHEMBL1172 |
| DrugBank | DB00967 |
| DrugCentral | 814 |
| FDA SRS | FVF865388R |
| Human Metabolome Database | HMDB0015102 |
| Guide to Pharmacology | 7157 |
| KEGG | D03693 |
| PharmGKB | PA164776964 |
| PubChem | 124087 |
| SureChEMBL | SCHEMBL4425 |
| ZINC | ZINC000000001261 |
 Cavia porcellus
Cavia porcellus
 Homo sapiens
Homo sapiens
 Mus musculus
Mus musculus