| Trade Names | |
| Synonyms | |
| Status | |
| Molecule Category | Salt-form |
| UNII | 08I79HTP27 |
| EPA CompTox | DTXSID4046024 |
| Parent Compound: | CLOPIDOGREL |
Structure
| InChI Key | FDEODCTUSIWGLK-RSAXXLAASA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C16H18ClNO6S2 |
| Molecular Weight | 419.91 |
| AlogP | 3.67 |
| Hydrogen Bond Acceptor | 4.0 |
| Number of Rotational Bond | 3.0 |
| Polar Surface Area | 29.54 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 2.0 |
| Heavy Atoms | 21.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Purinergic receptor P2Y12 antagonist | ANTAGONIST | DailyMed |
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Transporter
Electrochemical transporter
SLC superfamily of solute carriers
SLC21/SLCO family of organic anion transporting polypeptides
|
- | - | - | - | 96.14-145.29 |
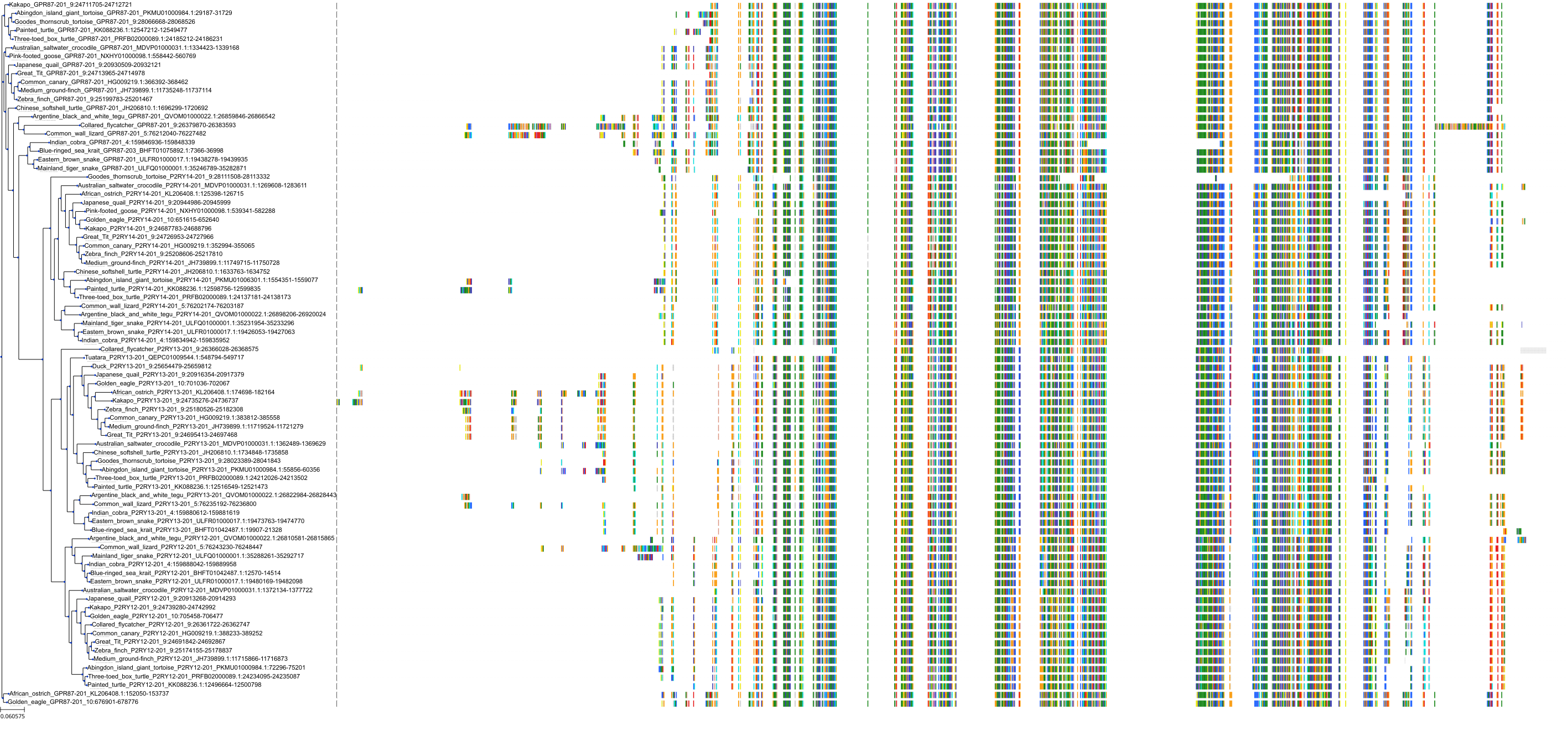
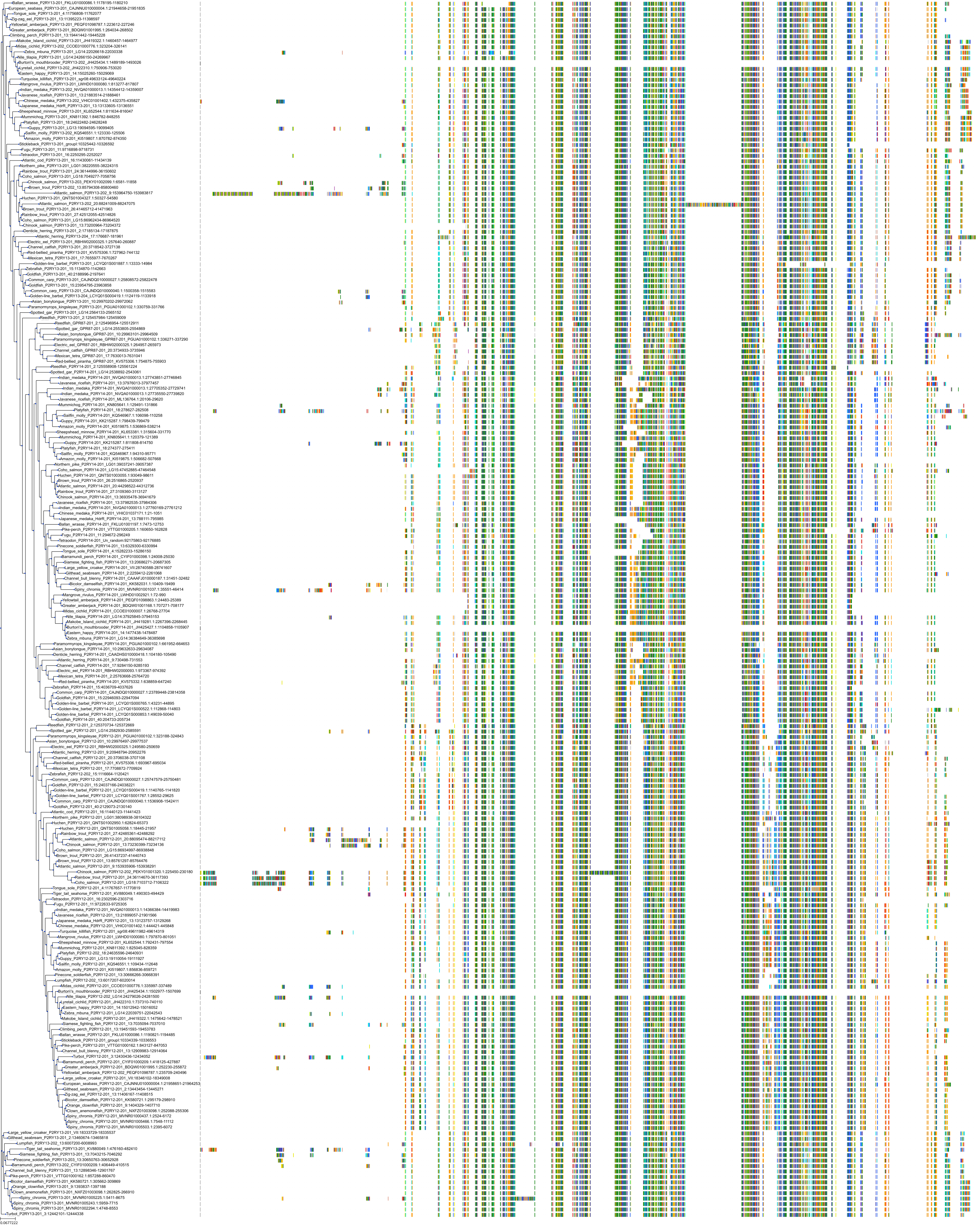
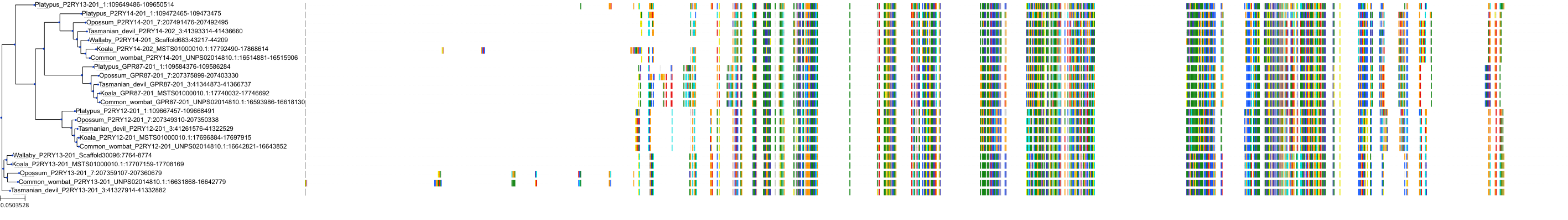
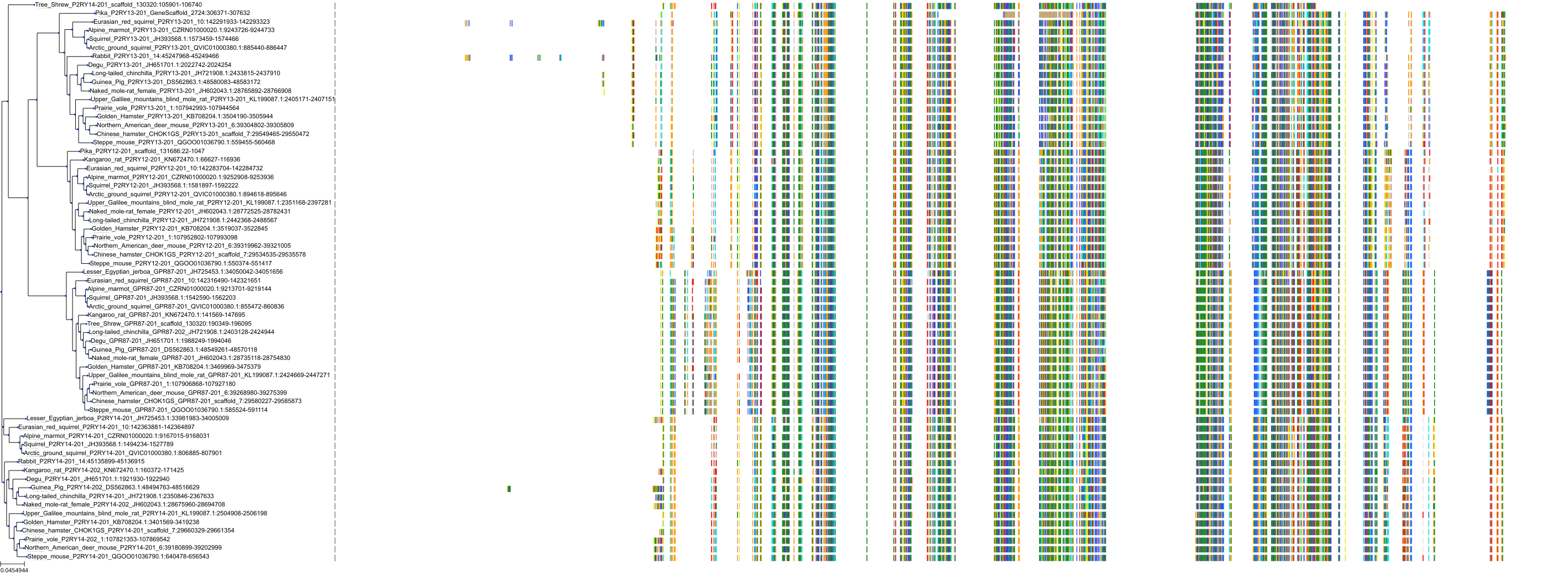
Target Conservation
|
Protein: Purinergic receptor P2Y12 Description: P2Y purinoceptor 12 Organism : Homo sapiens Q9H244 ENSG00000169313 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 3759 |
| ChEMBL | CHEMBL1083385 |
| FDA SRS | 08I79HTP27 |
| PubChem | 115366 |
| SureChEMBL | SCHEMBL33556 |
 Cricetulus griseus
Cricetulus griseus