| Trade Names | |
| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| UNII | 779619577M |
| EPA CompTox | DTXSID6045907 |
| Parent Compound: | CLOBETASOL |
Structure
| InChI Key | CBGUOGMQLZIXBE-XGQKBEPLSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C25H32ClFO5 |
| Molecular Weight | 466.98 |
| AlogP | 4.1 |
| Hydrogen Bond Acceptor | 5.0 |
| Hydrogen Bond Donor | 1.0 |
| Number of Rotational Bond | 4.0 |
| Polar Surface Area | 80.67 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 0.0 |
| Heavy Atoms | 32.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Glucocorticoid receptor agonist | AGONIST | PubMed |
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Enzyme
Cytochrome P450
Cytochrome P450 family 3
Cytochrome P450 family 3A
Cytochrome P450 3A4
|
- | 206 | - | - | - | |
|
Enzyme
Cytochrome P450
Cytochrome P450 family 3
Cytochrome P450 family 3A
Cytochrome P450 3A5
|
- | 21-103 | 100 | - | 90 | |
|
Transcription factor
Nuclear receptor
Nuclear hormone receptor subfamily 3
Nuclear hormone receptor subfamily 3 group C
Nuclear hormone receptor subfamily 3 group C member 1
|
- | 3.17 | - | - | - | |
|
Transporter
Electrochemical transporter
SLC superfamily of solute carriers
SLC21/SLCO family of organic anion transporting polypeptides
|
- | - | - | - | 20.25-21.34 |
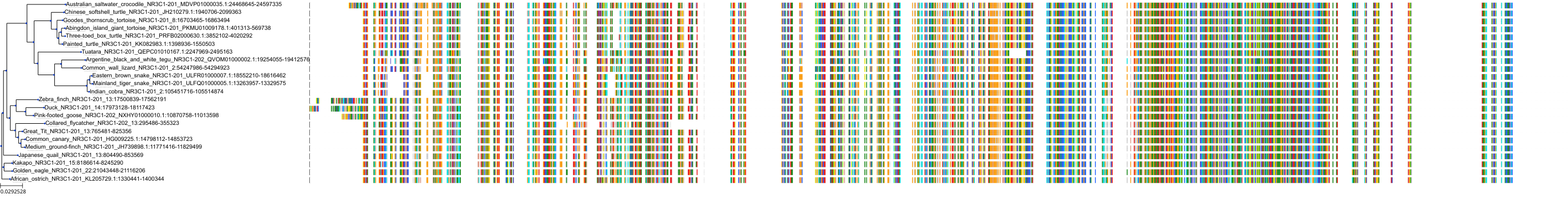
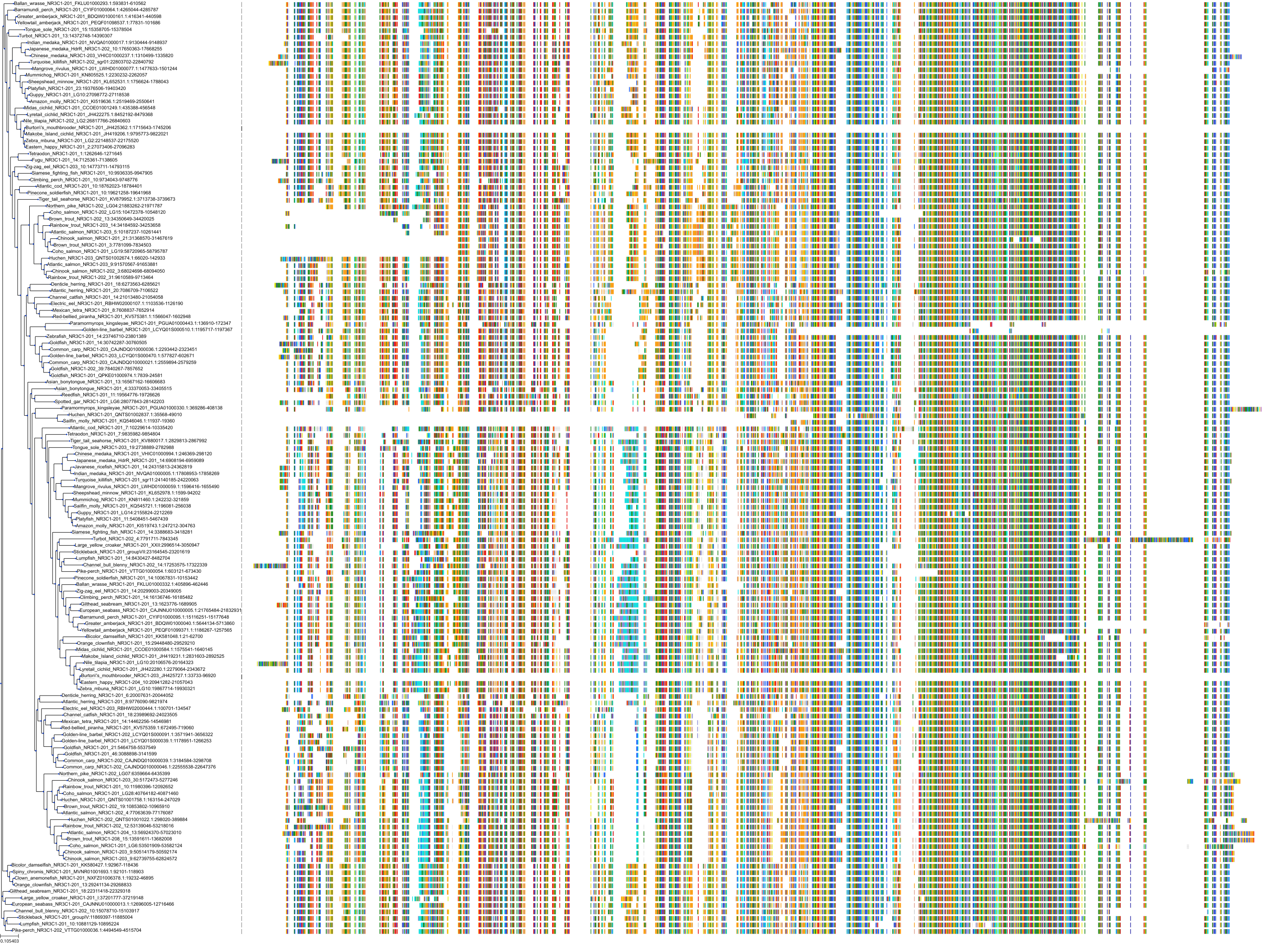
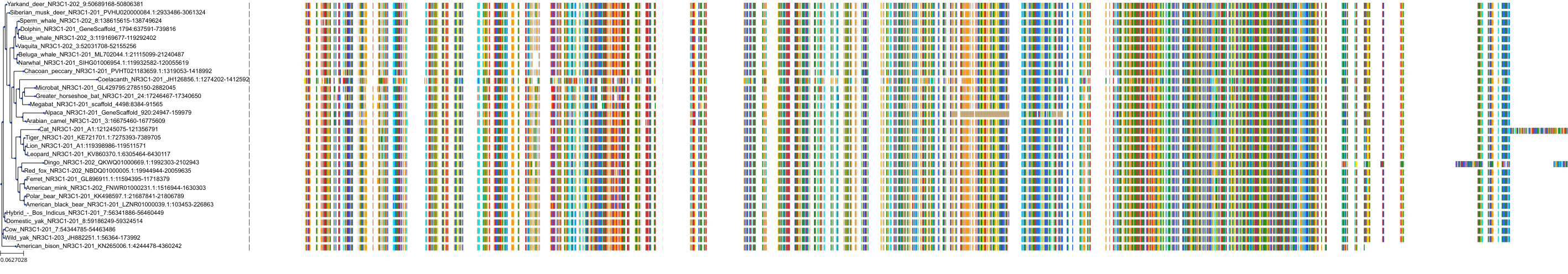
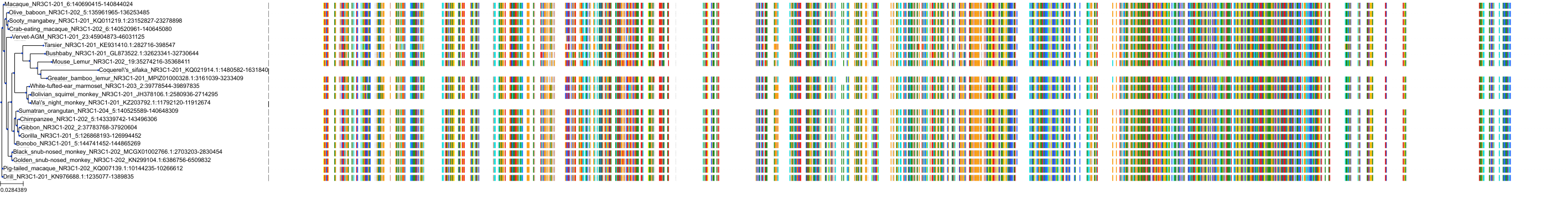
Target Conservation
|
Protein: Glucocorticoid receptor Description: Glucocorticoid receptor Organism : Homo sapiens P04150 ENSG00000113580 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 31414 |
| ChEMBL | CHEMBL1159650 |
| DrugBank | DB01013 |
| DrugCentral | 4452 |
| FDA SRS | 779619577M |
| Guide to Pharmacology | 7062 |
| PharmGKB | PA164744375 |
| PubChem | 32798 |
| SureChEMBL | SCHEMBL3997 |
| ZINC | ZINC000003977767 |
 Cricetulus griseus
Cricetulus griseus
 Homo sapiens
Homo sapiens
 Plasmodium falciparum
Plasmodium falciparum
 Rattus norvegicus
Rattus norvegicus