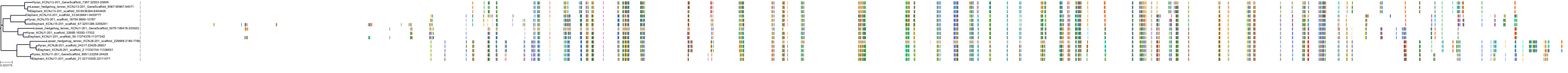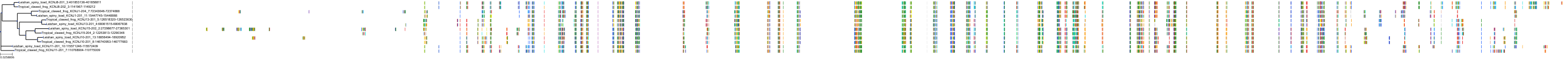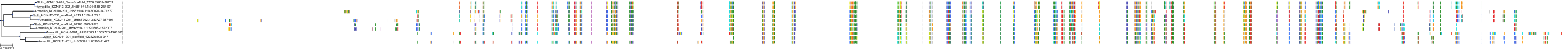| Trade Names | |
| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| ATC | A10BB02 |
| UNII | WTM2C3IL2X |
| EPA CompTox | DTXSID9020322 |
Structure
| InChI Key | RKWGIWYCVPQPMF-UHFFFAOYSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C10H13ClN2O3S |
| Molecular Weight | 276.75 |
| AlogP | 1.74 |
| Hydrogen Bond Acceptor | 3.0 |
| Hydrogen Bond Donor | 2.0 |
| Number of Rotational Bond | 4.0 |
| Polar Surface Area | 75.27 |
| Molecular species | ACID |
| Aromatic Rings | 1.0 |
| Heavy Atoms | 17.0 |
Metabolites Network
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Sulfonylurea receptor 1, Kir6.2 blocker | BLOCKER | PubMed PubMed Wikipedia FDA |
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Enzyme
Oxidoreductase
|
- | - | - | - | 0-0 | |
|
Transporter
Electrochemical transporter
SLC superfamily of solute carriers
SLC21/SLCO family of organic anion transporting polypeptides
|
- | - | - | - | 102.04-102.22 |
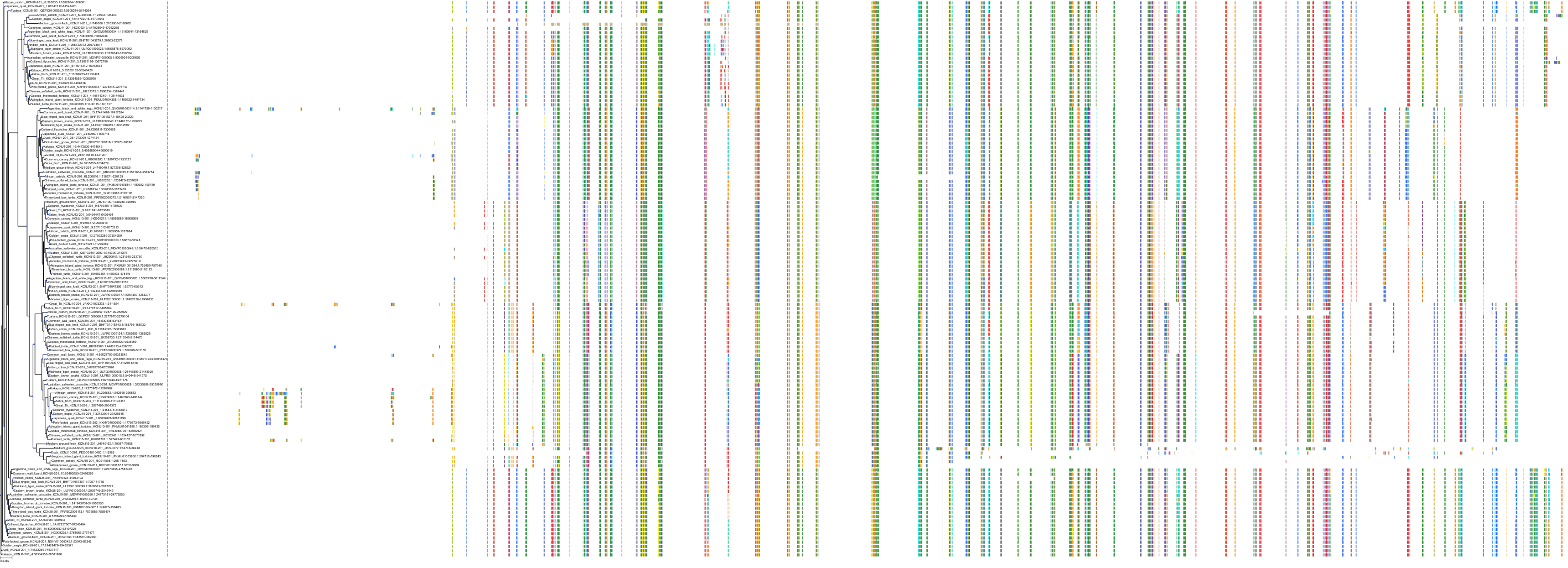
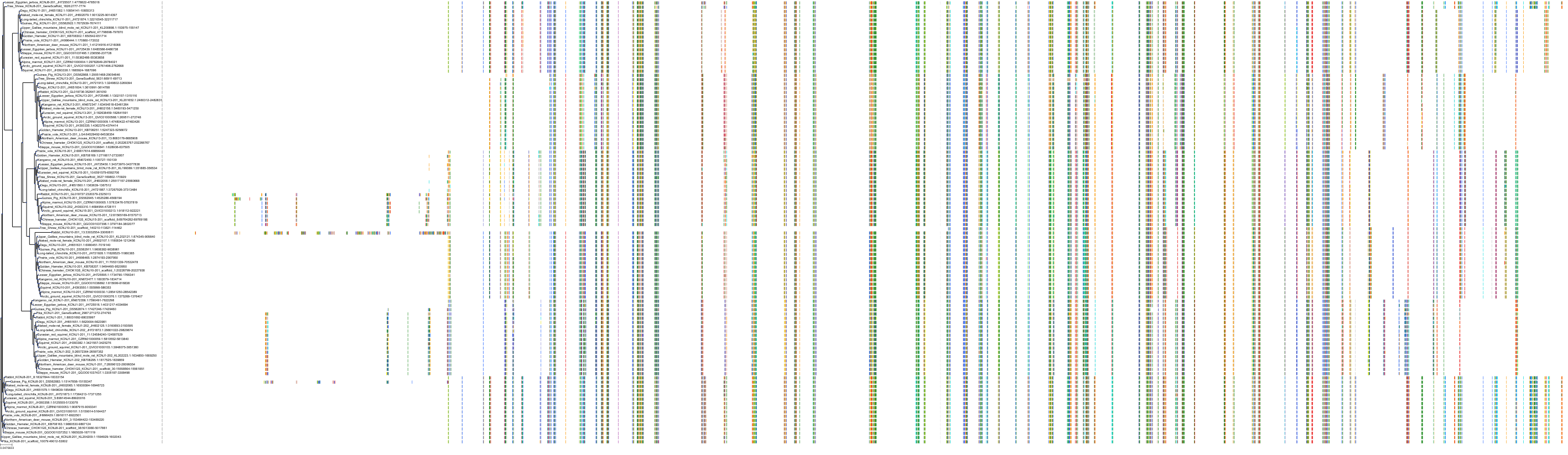
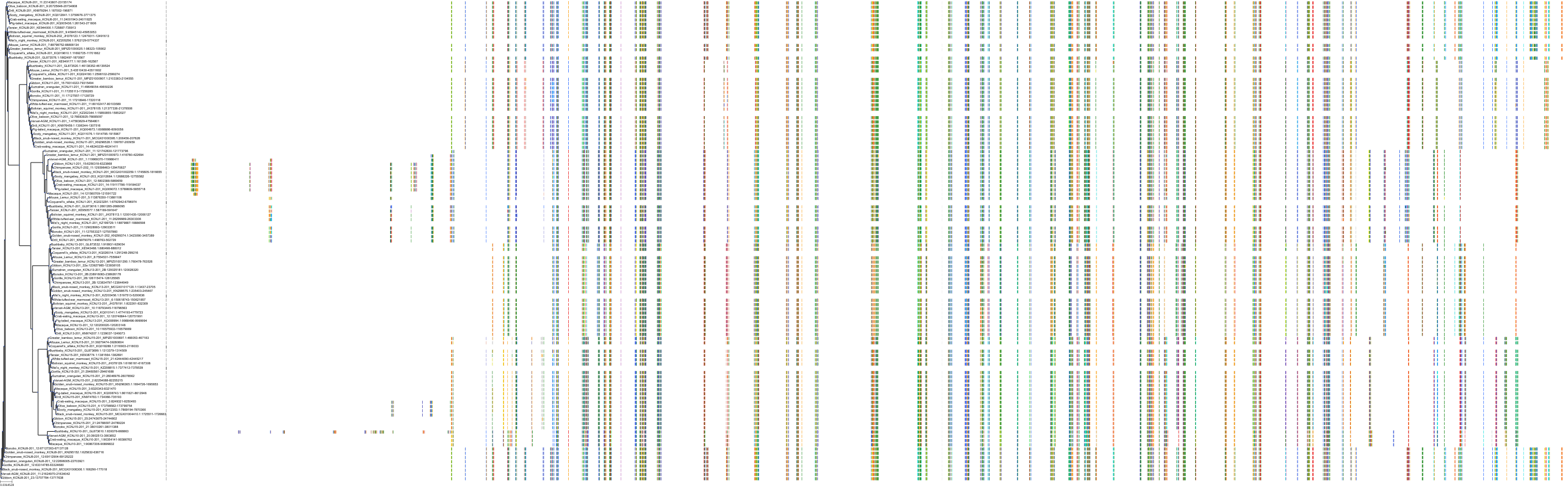
Target Conservation
|
Protein: Sulfonylurea receptor 1, Kir6.2 Description: ATP-binding cassette sub-family C member 8 Organism : Homo sapiens Q09428 ENSG00000006071 |
||||
|
Protein: Sulfonylurea receptor 1, Kir6.2 Description: ATP-sensitive inward rectifier potassium channel 11 Organism : Homo sapiens Q14654 ENSG00000187486 |
||||
Related Entries
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 3650 |
| ChEMBL | CHEMBL498 |
| DrugBank | DB00672 |
| DrugCentral | 622 |
| FDA SRS | WTM2C3IL2X |
| Human Metabolome Database | HMDB0014810 |
| Guide to Pharmacology | 6801 |
| KEGG | D00271 |
| PharmGKB | PA448966 |
| PubChem | 2727 |
| SureChEMBL | SCHEMBL23947 |
| ZINC | ZINC000001530599 |
 Cricetulus griseus
Cricetulus griseus
 Saccharomyces cerevisiae
Saccharomyces cerevisiae