| Trade Names | |
| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| ATC | C07AG02 |
| UNII | 0K47UL67F2 |
| EPA CompTox | DTXSID8022747 |
Structure
| InChI Key | OGHNVEJMJSYVRP-UHFFFAOYSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C24H26N2O4 |
| Molecular Weight | 406.48 |
| AlogP | 3.74 |
| Hydrogen Bond Acceptor | 5.0 |
| Hydrogen Bond Donor | 3.0 |
| Number of Rotational Bond | 10.0 |
| Polar Surface Area | 75.74 |
| Molecular species | BASE |
| Aromatic Rings | 4.0 |
| Heavy Atoms | 30.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Adrenergic receptor alpha-1 antagonist | ANTAGONIST | FDA |
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Ion channel
Voltage-gated ion channel
Potassium channels
Voltage-gated potassium channel
|
- | 350 | - | - | - | |
|
Membrane receptor
Family A G protein-coupled receptor
Small molecule receptor (family A GPCR)
Monoamine receptor
Adrenergic receptor
|
- | - | 0.195 | 0.81-3.4 | - | |
|
Membrane receptor
Family A G protein-coupled receptor
Small molecule receptor (family A GPCR)
Monoamine receptor
Serotonin receptor
|
- | - | - | 3.162 | - | |
|
Transporter
Electrochemical transporter
SLC superfamily of solute carriers
SLC47 family of multidrug and toxin extrusion transporters
|
- | - | - | - | 21 |
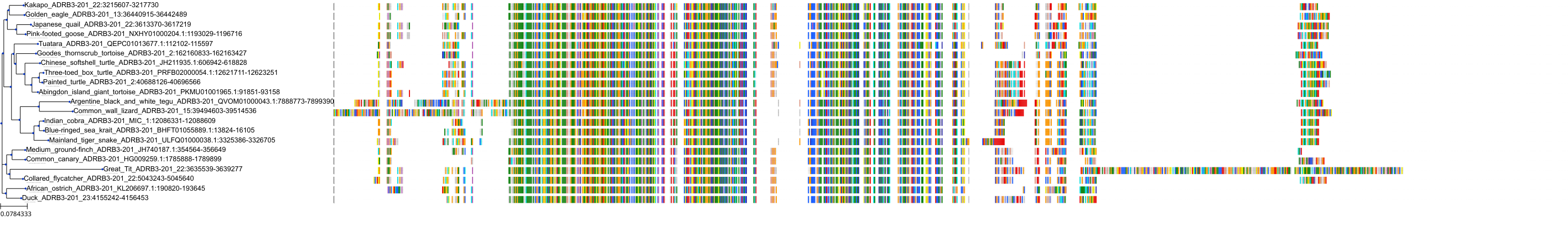
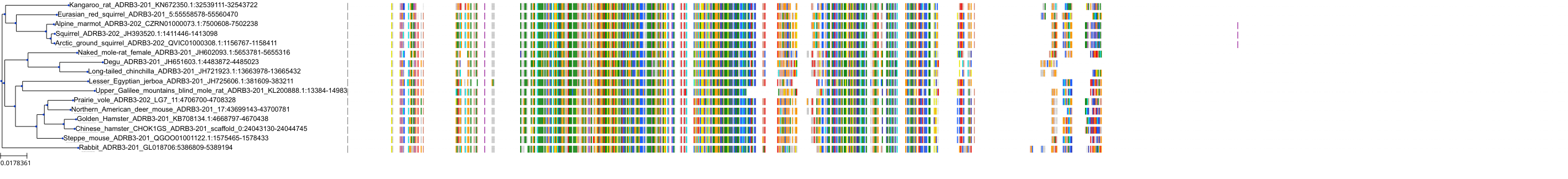
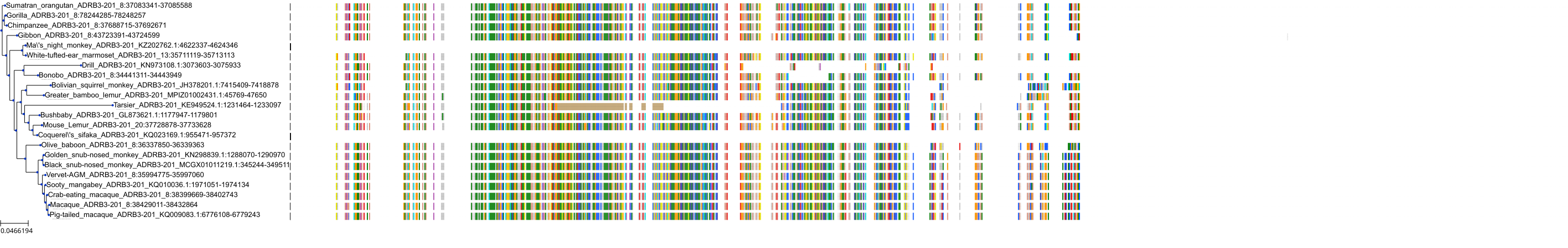
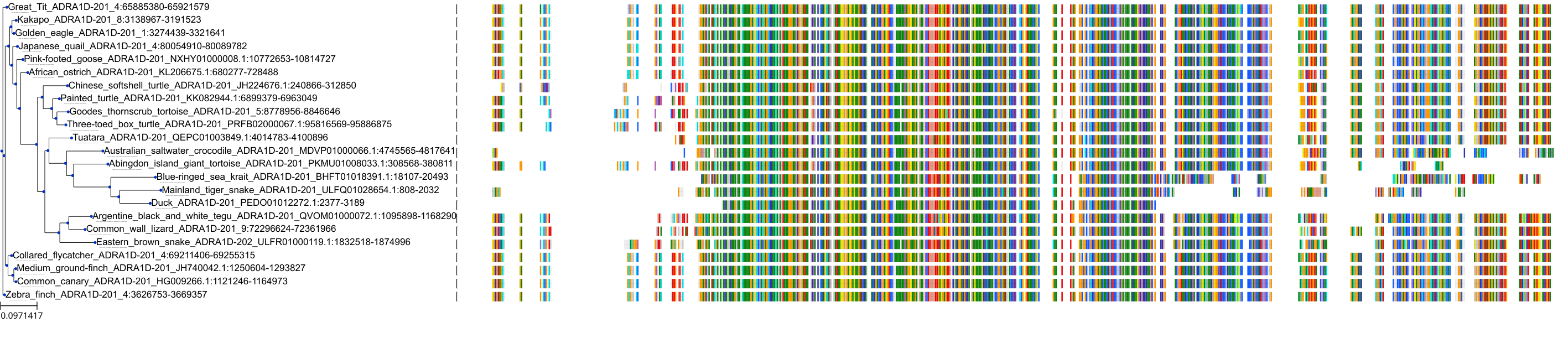
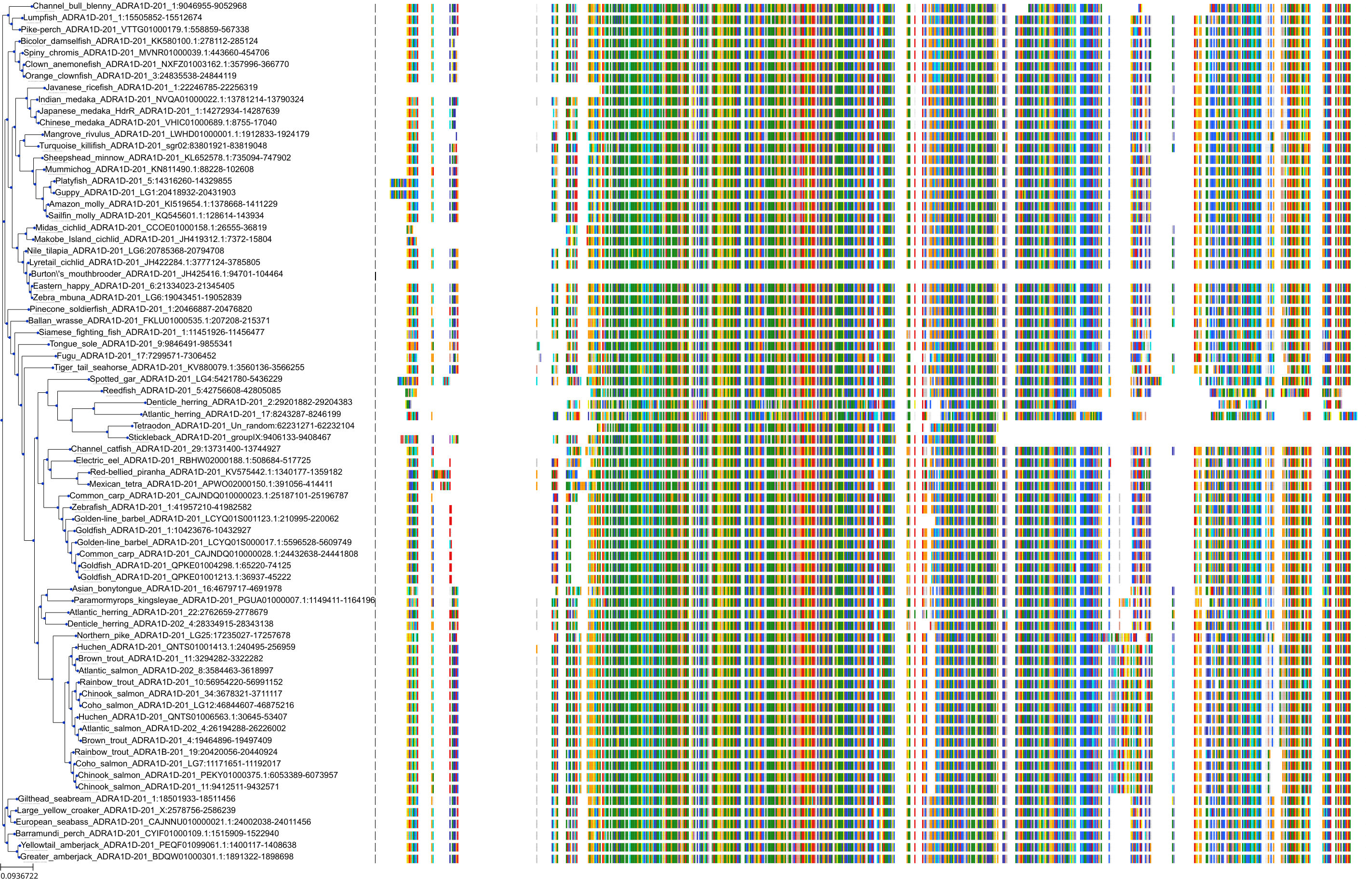
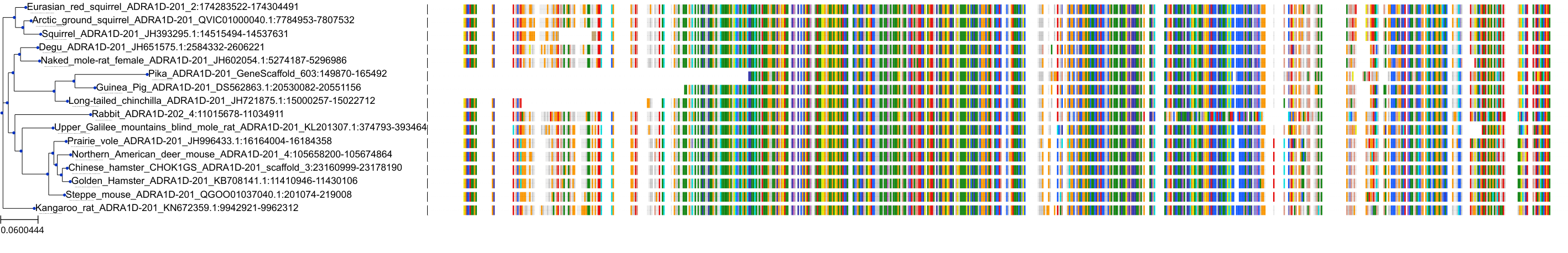
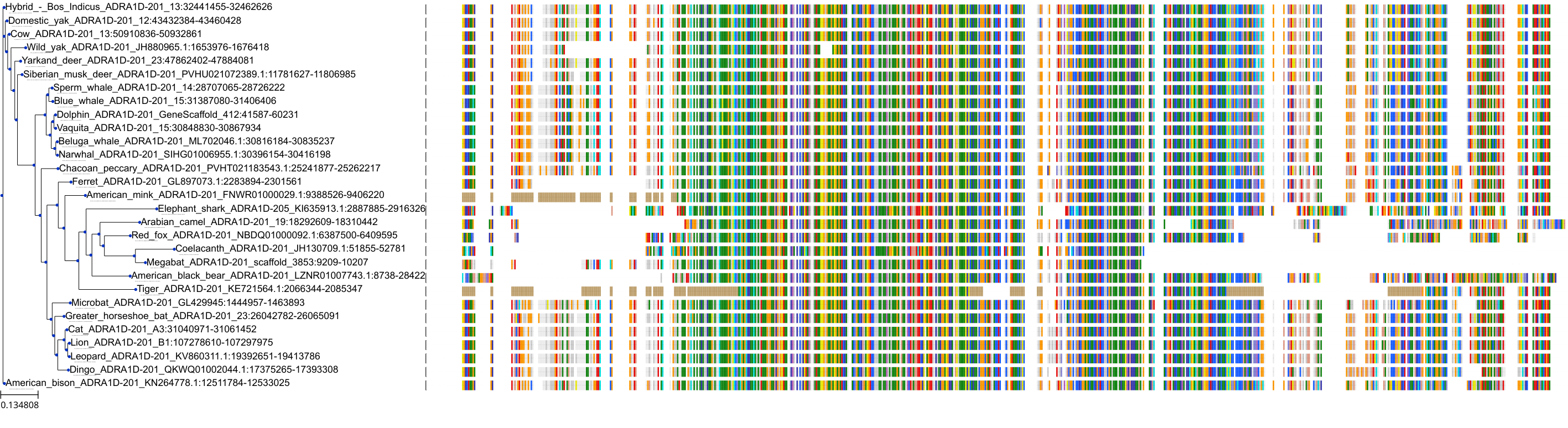
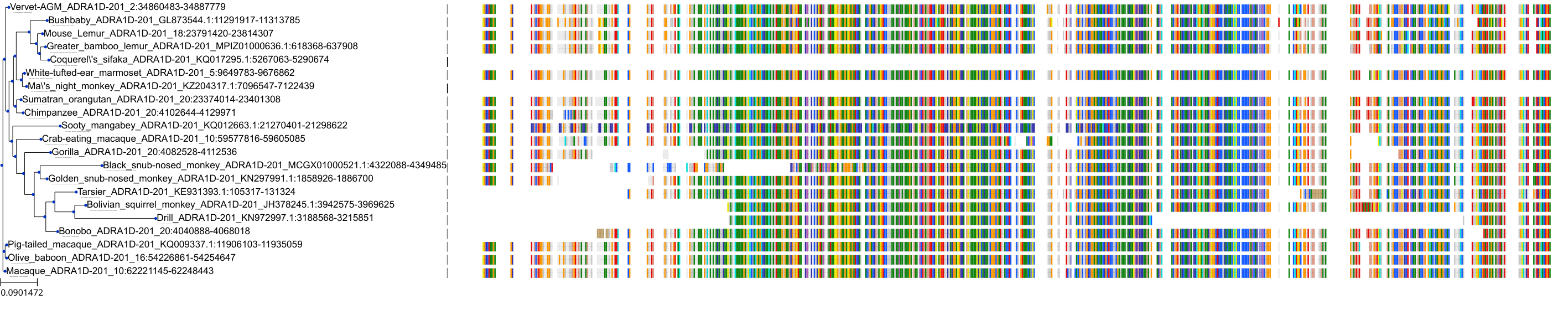
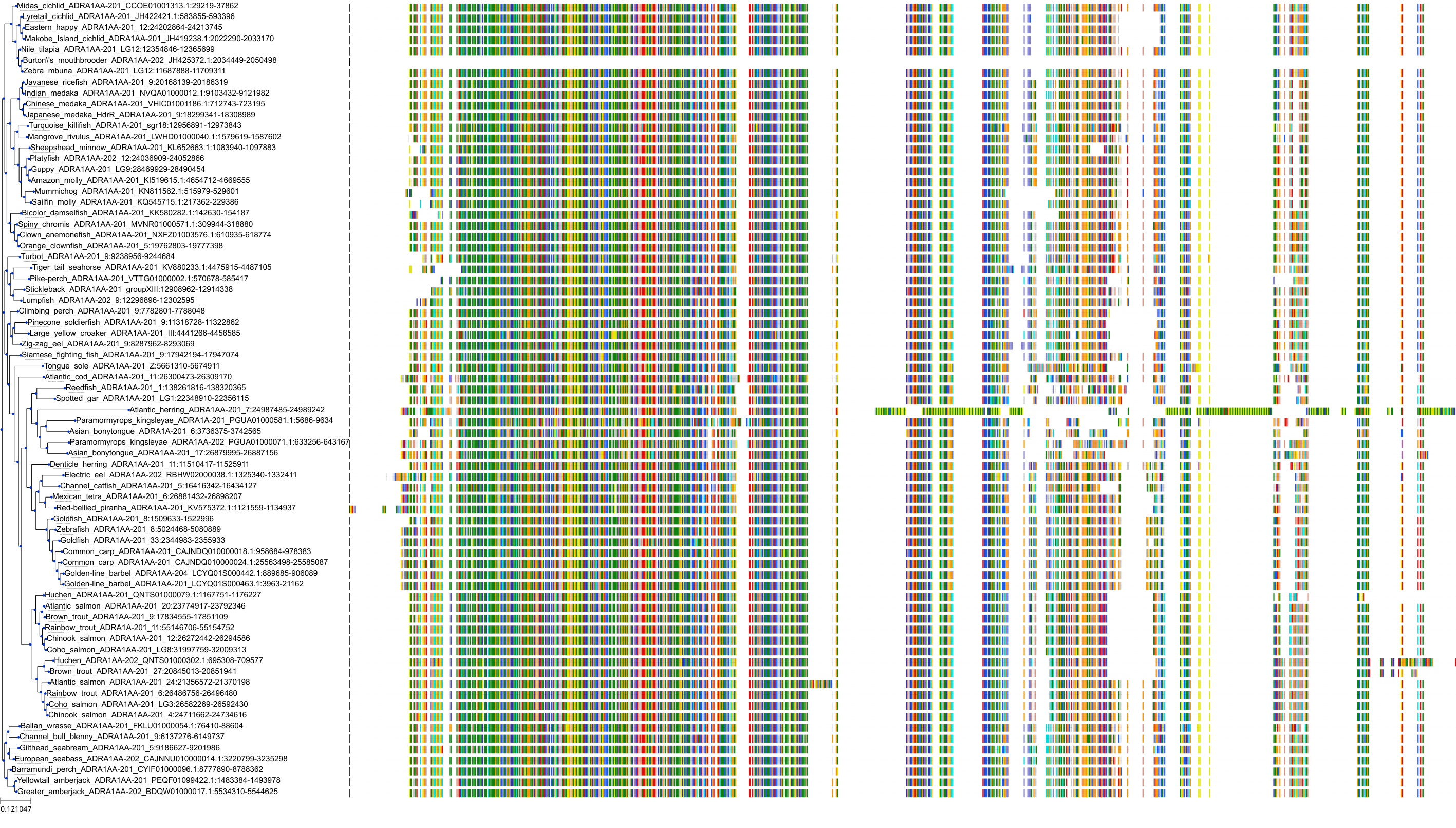
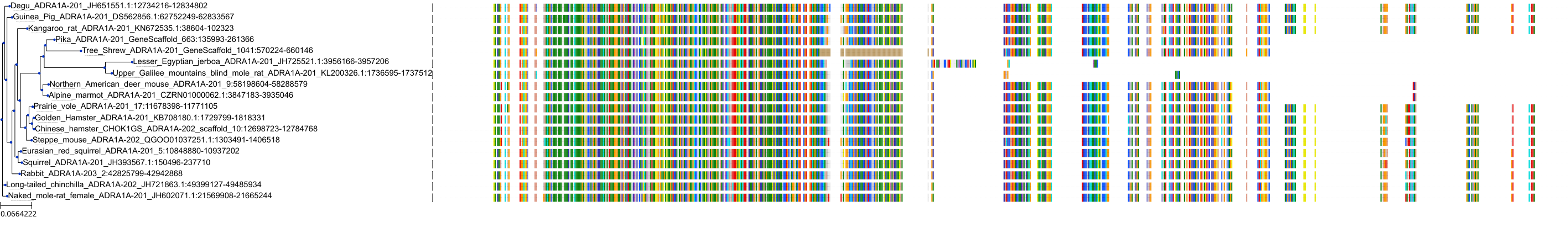
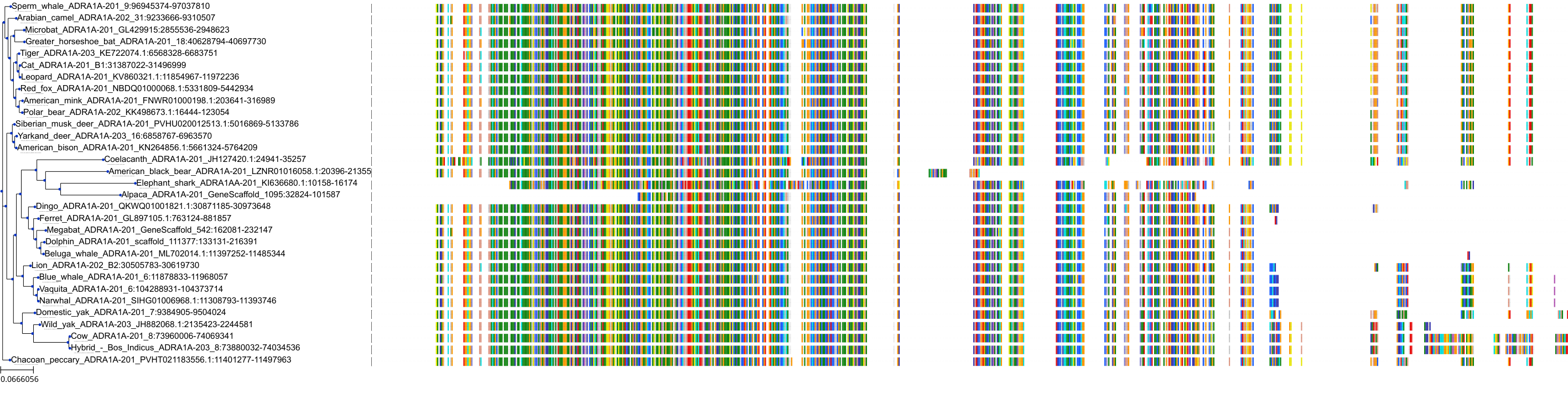
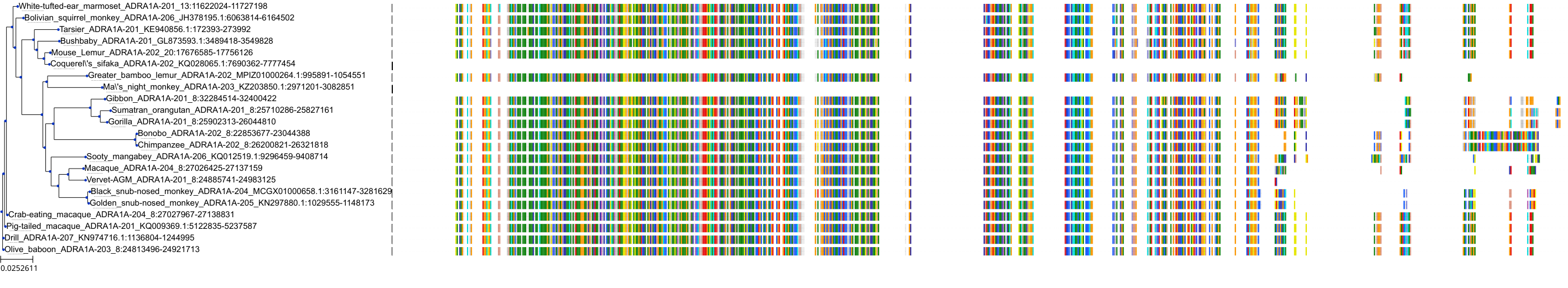
Target Conservation
|
Protein: Adrenergic receptor beta Description: Beta-2 adrenergic receptor Organism : Homo sapiens P07550 ENSG00000169252 |
||||
|
Protein: Adrenergic receptor beta Description: Beta-1 adrenergic receptor Organism : Homo sapiens P08588 ENSG00000043591 |
||||
|
Protein: Adrenergic receptor beta Description: Beta-3 adrenergic receptor Organism : Homo sapiens P13945 ENSG00000188778 |
||||
|
Protein: Adrenergic receptor alpha-1 Description: Alpha-1D adrenergic receptor Organism : Homo sapiens P25100 ENSG00000171873 |
||||
|
Protein: Adrenergic receptor alpha-1 Description: Alpha-1A adrenergic receptor Organism : Homo sapiens P35348 ENSG00000120907 |
||||
|
Protein: Adrenergic receptor alpha-1 Description: Alpha-1B adrenergic receptor Organism : Homo sapiens P35368 ENSG00000170214 |
||||
Related Entries
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 3441 |
| ChEMBL | CHEMBL723 |
| DrugBank | DB01136 |
| DrugCentral | 522 |
| FDA SRS | 0K47UL67F2 |
| Human Metabolome Database | HMDB0015267 |
| Guide to Pharmacology | 551 |
| KEGG | C06875 |
| PharmGKB | PA448817 |
| PubChem | 2585 |
| SureChEMBL | SCHEMBL22293 |
| ZINC | ZINC01530580 |
 Homo sapiens
Homo sapiens
 Mus musculus
Mus musculus
 Rattus norvegicus
Rattus norvegicus