| Trade Names | |
| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| ATC | L01EE03 |
| UNII | 181R97MR71 |
| EPA CompTox | DTXSID70209422 |
Structure
| InChI Key | ACWZRVQXLIRSDF-UHFFFAOYSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C17H15BrF2N4O3 |
| Molecular Weight | 441.23 |
| AlogP | 3.01 |
| Hydrogen Bond Acceptor | 6.0 |
| Hydrogen Bond Donor | 3.0 |
| Number of Rotational Bond | 6.0 |
| Polar Surface Area | 88.41 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 3.0 |
| Heavy Atoms | 27.0 |
Pharmacology
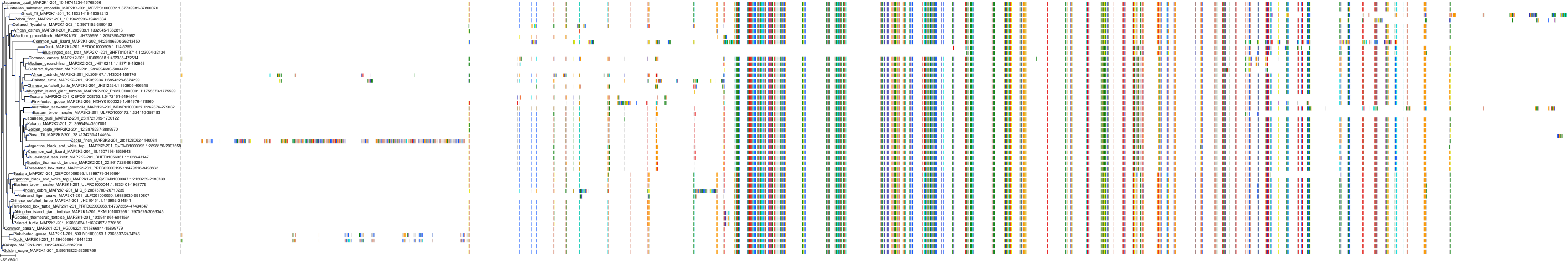
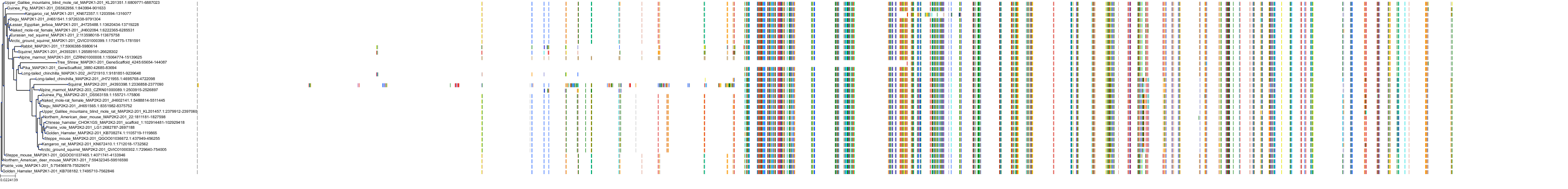
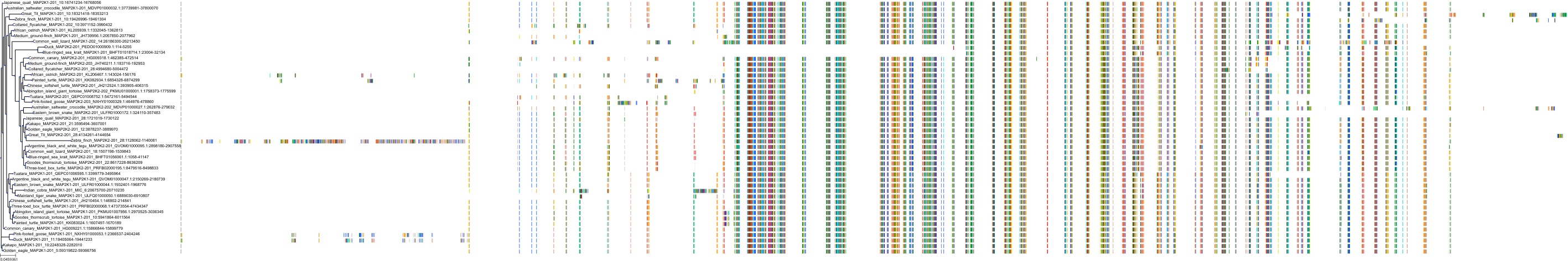
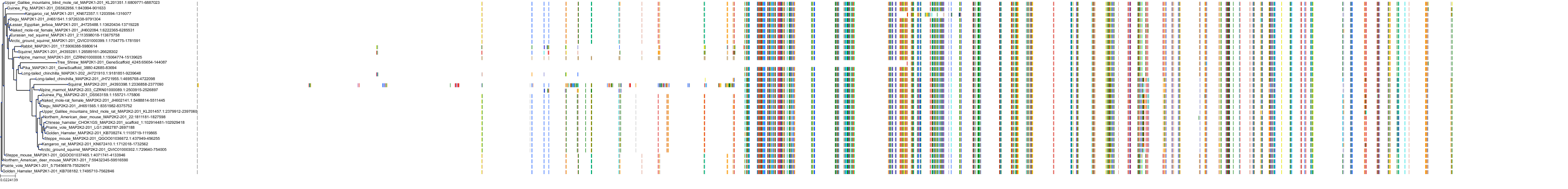
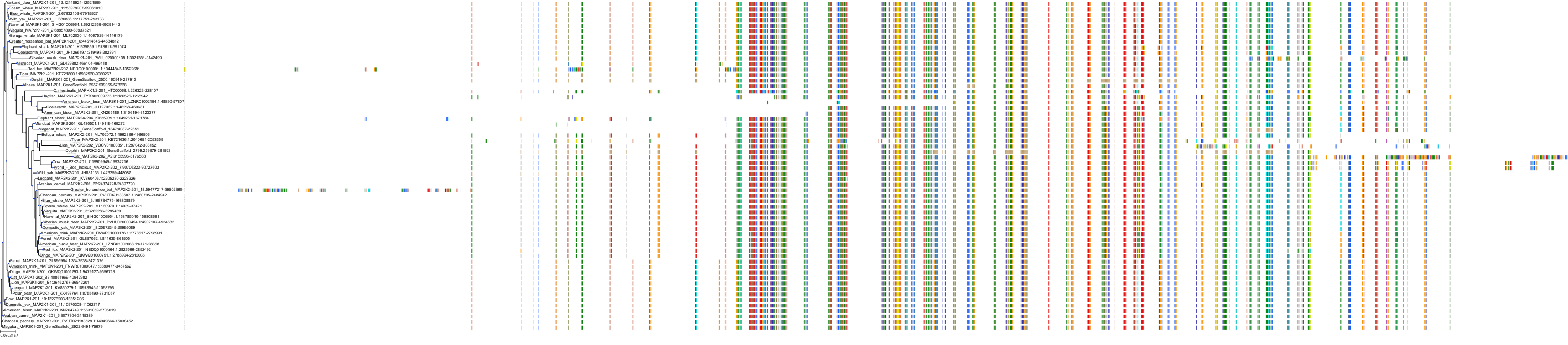
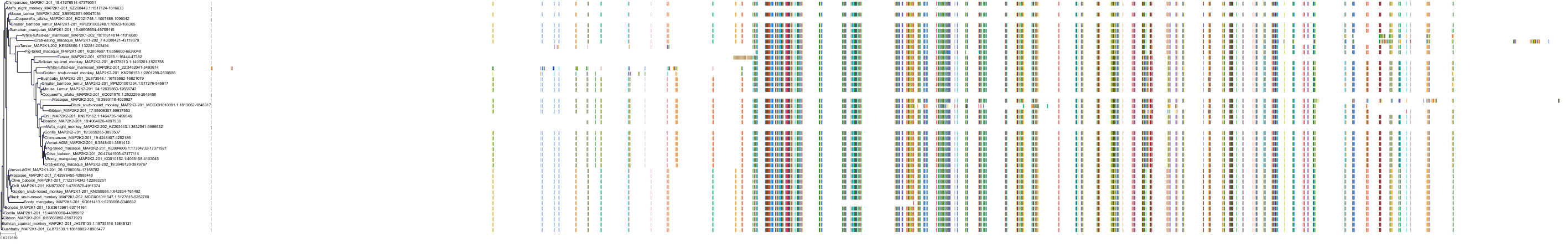
Target Conservation
|
Protein: Dual specificity mitogen-activated protein kinase kinase 2 Description: Dual specificity mitogen-activated protein kinase kinase 2 Organism : Homo sapiens P36507 ENSG00000126934 |
||||
|
Protein: Dual specificity mitogen-activated protein kinase kinase 1 Description: Dual specificity mitogen-activated protein kinase kinase 1 Organism : Homo sapiens Q02750 ENSG00000169032 |
||||
Related Entries
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 145371 |
| ChEMBL | CHEMBL3187723 |
| DrugBank | DB11967 |
| DrugCentral | 5290 |
| FDA SRS | 181R97MR71 |
| Guide to Pharmacology | 7921 |
| PDB | QO7 |
| PharmGKB | PA166179867 |
| PubChem | 10288191 |
| SureChEMBL | SCHEMBL570088 |
| ZINC | ZINC000038460704 |
 Homo sapiens
Homo sapiens