| Trade Names | |
| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| ATC | G04BE10 |
| UNII | DR5S136IVO |
| EPA CompTox | DTXSID50186727 |
Structure
| InChI Key | WEAJZXNPAWBCOA-INIZCTEOSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C23H26ClN7O3 |
| Molecular Weight | 483.96 |
| AlogP | 1.85 |
| Hydrogen Bond Acceptor | 8.0 |
| Hydrogen Bond Donor | 3.0 |
| Number of Rotational Bond | 8.0 |
| Polar Surface Area | 128.62 |
| Molecular species | BASE |
| Aromatic Rings | 3.0 |
| Heavy Atoms | 34.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Phosphodiesterase 5A inhibitor | INHIBITOR | FDA |
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Enzyme
Phosphodiesterase
Phosphodiesterase 5
Phosphodiesterase 5A
|
- | 5.2 | - | 5 | 66-84 |
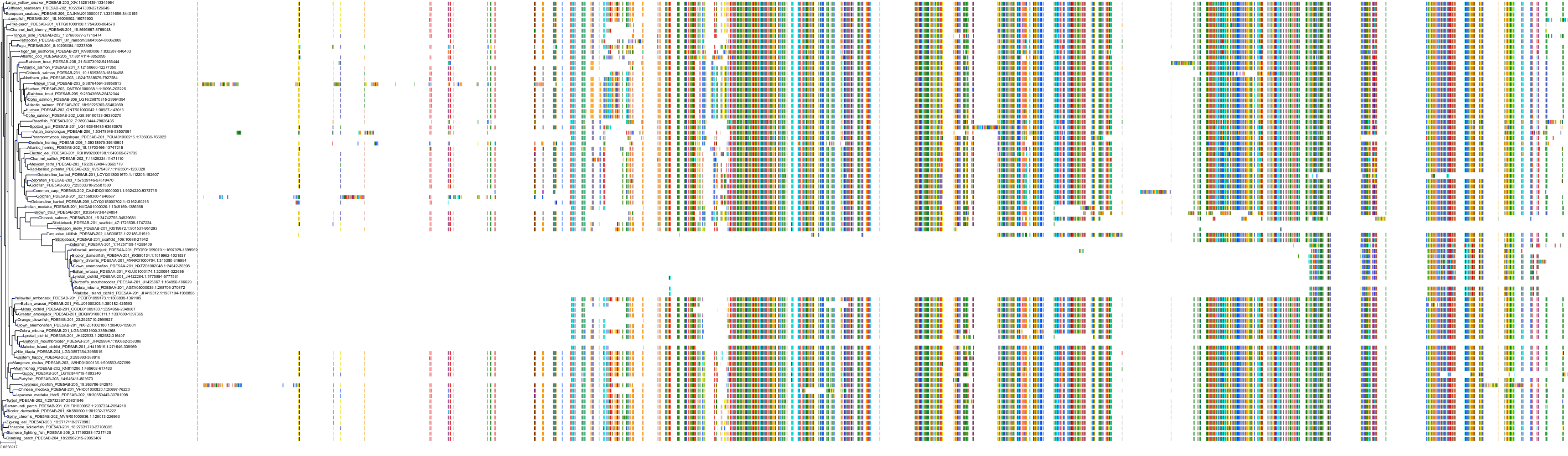
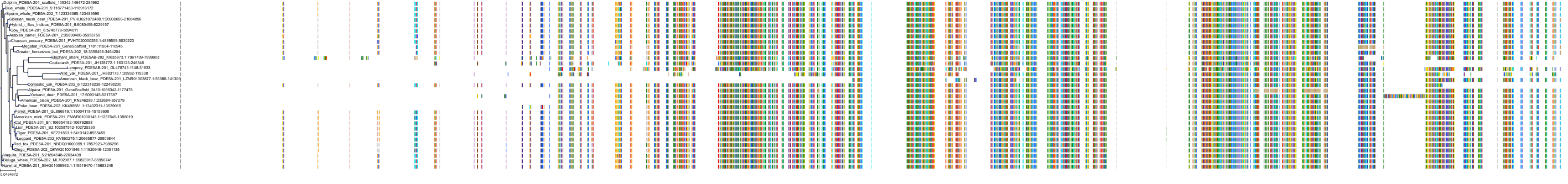
Target Conservation
|
Protein: Phosphodiesterase 5A Description: cGMP-specific 3',5'-cyclic phosphodiesterase Organism : Homo sapiens O76074 ENSG00000138735 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 66876 |
| ChEMBL | CHEMBL1963681 |
| DrugBank | DB06237 |
| DrugCentral | 4305 |
| FDA SRS | DR5S136IVO |
| Guide to Pharmacology | 7448 |
| PDB | E6L |
| PubChem | 9869929 |
| SureChEMBL | SCHEMBL118799 |
| ZINC | ZINC000011677857 |
 Canis lupus familiaris
Canis lupus familiaris
 Homo sapiens
Homo sapiens