| Trade Names | |
| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| ATC | N06AA17 |
| UNII | R63VQ857OT |
| EPA CompTox | DTXSID7022598 |
Structure
| InChI Key | QWGDMFLQWFTERH-UHFFFAOYSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C17H16ClN3O |
| Molecular Weight | 313.79 |
| AlogP | 3.43 |
| Hydrogen Bond Acceptor | 4.0 |
| Hydrogen Bond Donor | 1.0 |
| Polar Surface Area | 36.86 |
| Molecular species | BASE |
| Aromatic Rings | 2.0 |
| Heavy Atoms | 22.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Dopamine receptor antagonist | ANTAGONIST | DailyMed |
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Enzyme
Hydrolase
|
- | - | - | - | 7.3 | |
|
Membrane receptor
Family A G protein-coupled receptor
Small molecule receptor (family A GPCR)
Monoamine receptor
Adrenergic receptor
|
- | - | - | - | 43 | |
|
Membrane receptor
Family A G protein-coupled receptor
Small molecule receptor (family A GPCR)
Monoamine receptor
Dopamine receptor
|
- | - | - | 34 | - | |
|
Membrane receptor
Family A G protein-coupled receptor
Small molecule receptor (family A GPCR)
Monoamine receptor
Serotonin receptor
|
- | - | - | 1.77-500 | - | |
|
Transporter
Electrochemical transporter
SLC superfamily of solute carriers
SLC21/SLCO family of organic anion transporting polypeptides
|
- | - | - | - | 101.48-115.83 |
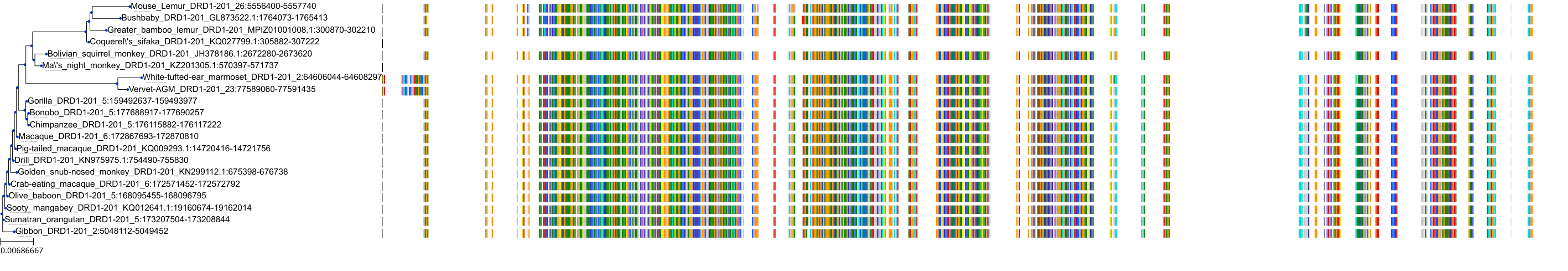
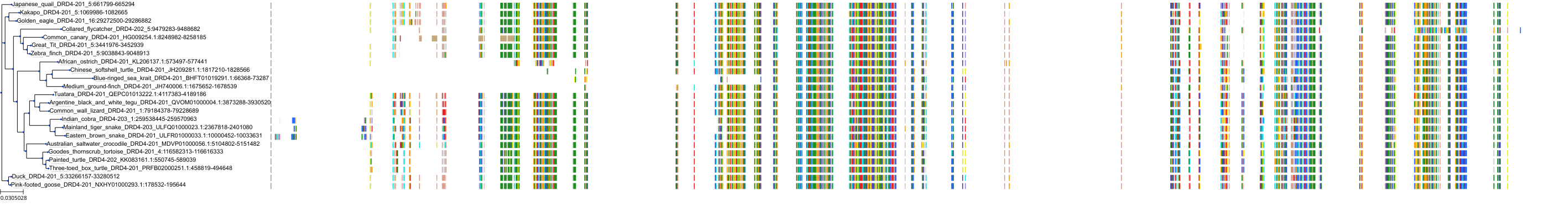
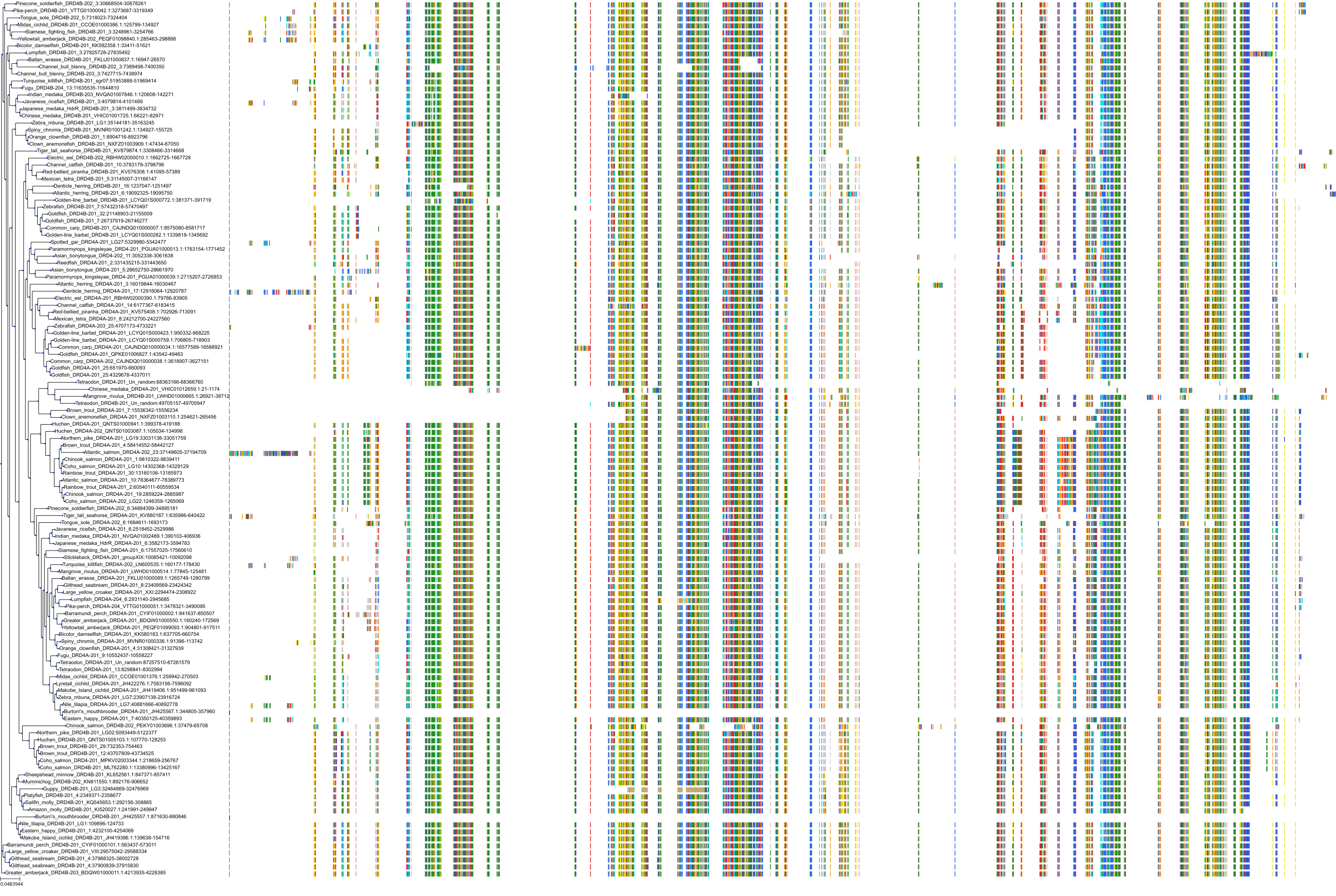
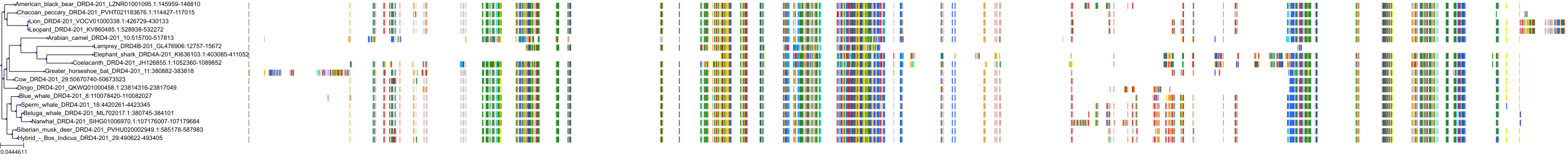
Target Conservation
|
Protein: Dopamine receptor Description: D(2) dopamine receptor Organism : Homo sapiens P14416 ENSG00000149295 |
||||
|
Protein: Dopamine receptor Description: D(1A) dopamine receptor Organism : Homo sapiens P21728 ENSG00000184845 |
||||
|
Protein: Dopamine receptor Description: D(4) dopamine receptor Organism : Homo sapiens P21917 ENSG00000069696 |
||||
|
Protein: Dopamine receptor Description: D(1B) dopamine receptor Organism : Homo sapiens P21918 ENSG00000169676 |
||||
|
Protein: Norepinephrine transporter Description: Sodium-dependent noradrenaline transporter Organism : Homo sapiens P23975 ENSG00000103546 |
||||
|
Protein: Serotonin transporter Description: Sodium-dependent serotonin transporter Organism : Homo sapiens P31645 ENSG00000108576 |
||||
|
Protein: Dopamine receptor Description: D(3) dopamine receptor Organism : Homo sapiens P35462 ENSG00000151577 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 2675 |
| ChEMBL | CHEMBL1113 |
| DrugBank | DB00543 |
| DrugCentral | 191 |
| FDA SRS | R63VQ857OT |
| Human Metabolome Database | HMDB0014683 |
| Guide to Pharmacology | 201 |
| KEGG | D00228 |
| PharmGKB | PA448405 |
| PubChem | 2170 |
| SureChEMBL | SCHEMBL33950 |
| ZINC | ZINC000000000931 |
 Bacteria
Bacteria
 Cricetulus griseus
Cricetulus griseus
 Escherichia coli
Escherichia coli
 Homo sapiens
Homo sapiens