| Trade Names | |
| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| ATC | D10AD03 |
| UNII | 1L4806J2QF |
| EPA CompTox | DTXSID5046481 |
Structure
| InChI Key | LZCDAPDGXCYOEH-UHFFFAOYSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C28H28O3 |
| Molecular Weight | 412.53 |
| AlogP | 6.68 |
| Hydrogen Bond Acceptor | 2.0 |
| Hydrogen Bond Donor | 1.0 |
| Number of Rotational Bond | 4.0 |
| Polar Surface Area | 46.53 |
| Molecular species | ACID |
| Aromatic Rings | 3.0 |
| Heavy Atoms | 31.0 |
Pharmacology
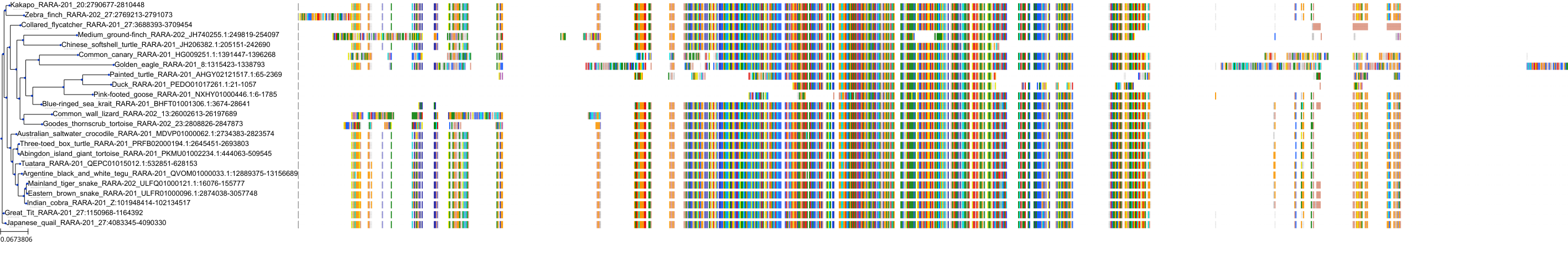
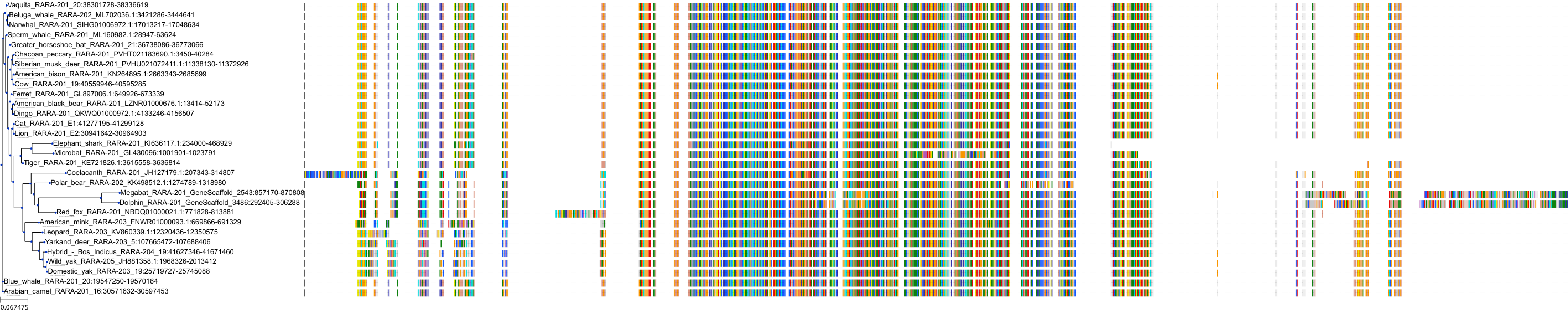
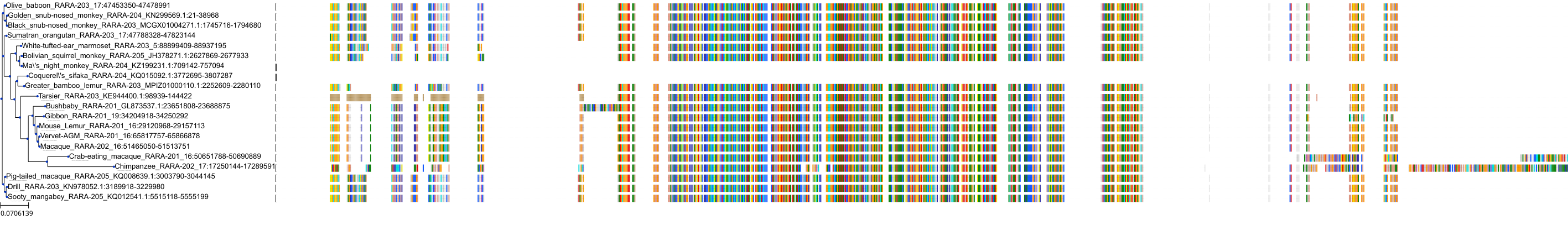
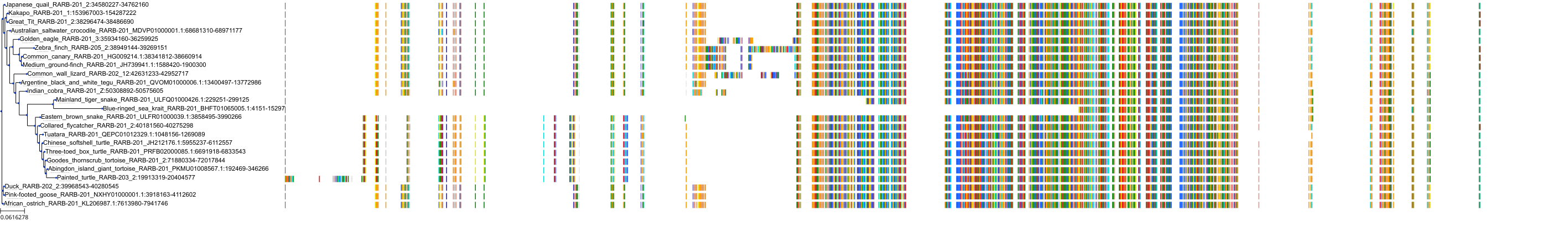
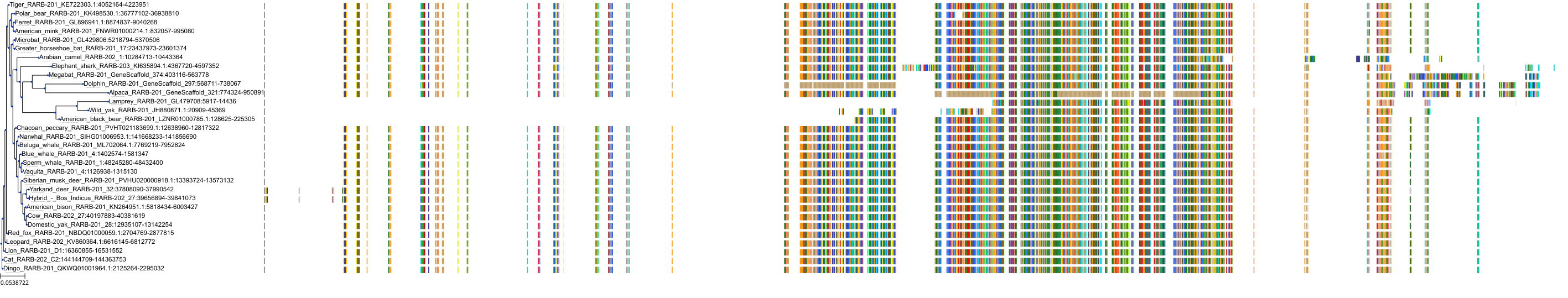
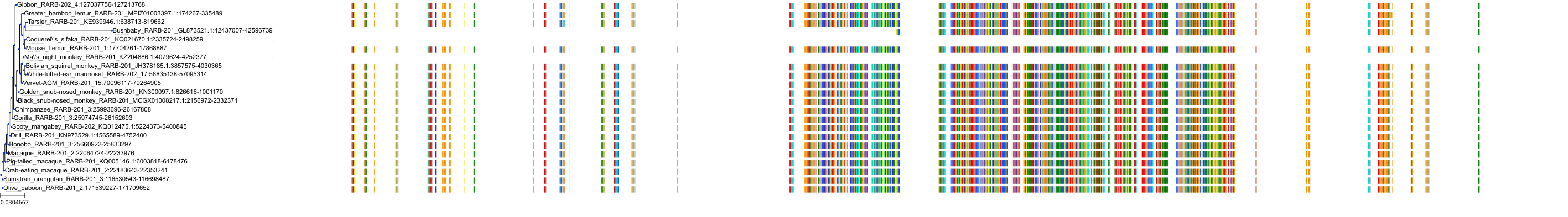
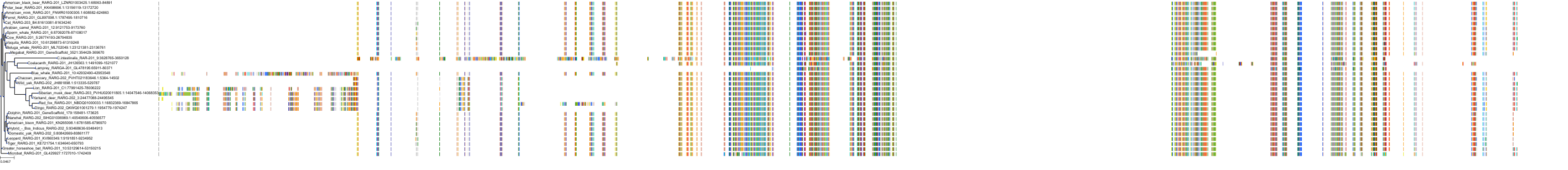
Target Conservation
|
Protein: Retinoic acid receptor Description: Retinoic acid receptor alpha Organism : Homo sapiens P10276 ENSG00000131759 |
||||
|
Protein: Retinoic acid receptor Description: Retinoic acid receptor beta Organism : Homo sapiens P10826 ENSG00000077092 |
||||
|
Protein: Retinoic acid receptor Description: Retinoic acid receptor gamma Organism : Homo sapiens P13631 ENSG00000172819 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 31174 |
| ChEMBL | CHEMBL1265 |
| DrugBank | DB00210 |
| DrugCentral | 87 |
| FDA SRS | 1L4806J2QF |
| Human Metabolome Database | HMDB0014355 |
| Guide to Pharmacology | 5429 |
| KEGG | D01112 |
| PharmGKB | PA448047 |
| PubChem | 60164 |
| SureChEMBL | SCHEMBL2747 |
| ZINC | ZINC000003784182 |
 Escherichia coli
Escherichia coli
 Homo sapiens
Homo sapiens