| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| ATC | R06AX29 |
| UNII | PA1123N395 |
| EPA CompTox | DTXSID5057678 |
Structure
| InChI Key | ACCMWZWAEFYUGZ-UHFFFAOYSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C28H37N3O3 |
| Molecular Weight | 463.62 |
| AlogP | 4.86 |
| Hydrogen Bond Acceptor | 5.0 |
| Hydrogen Bond Donor | 1.0 |
| Number of Rotational Bond | 10.0 |
| Polar Surface Area | 67.59 |
| Molecular species | ZWITTERION |
| Aromatic Rings | 3.0 |
| Heavy Atoms | 34.0 |
Pharmacology
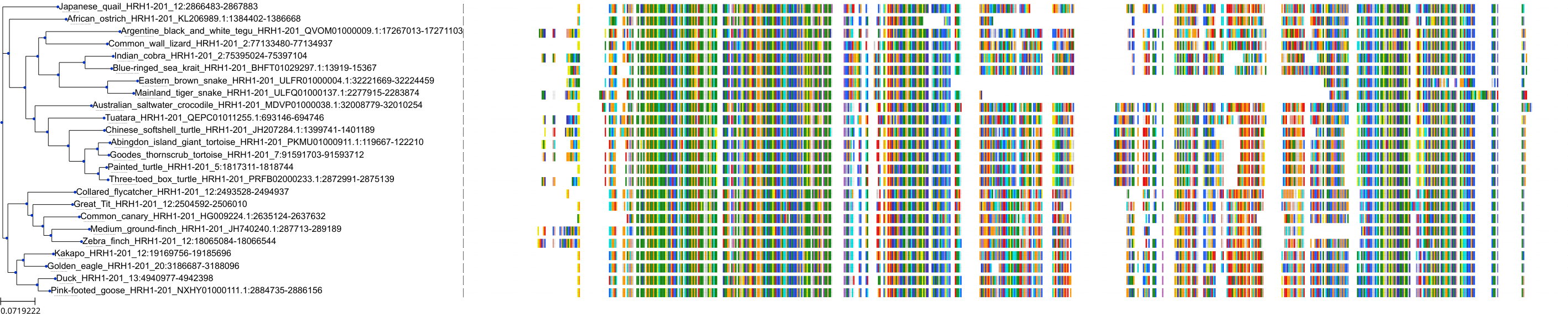
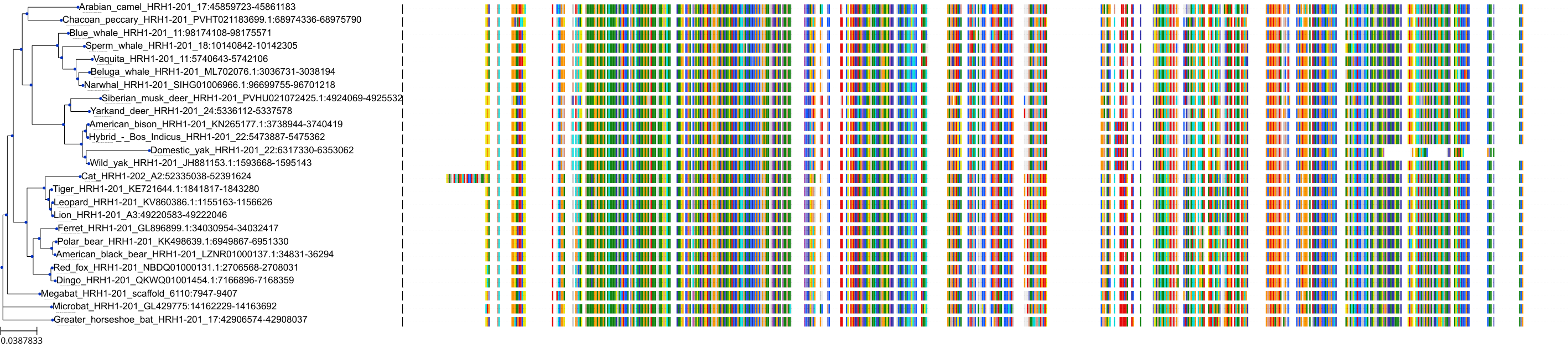
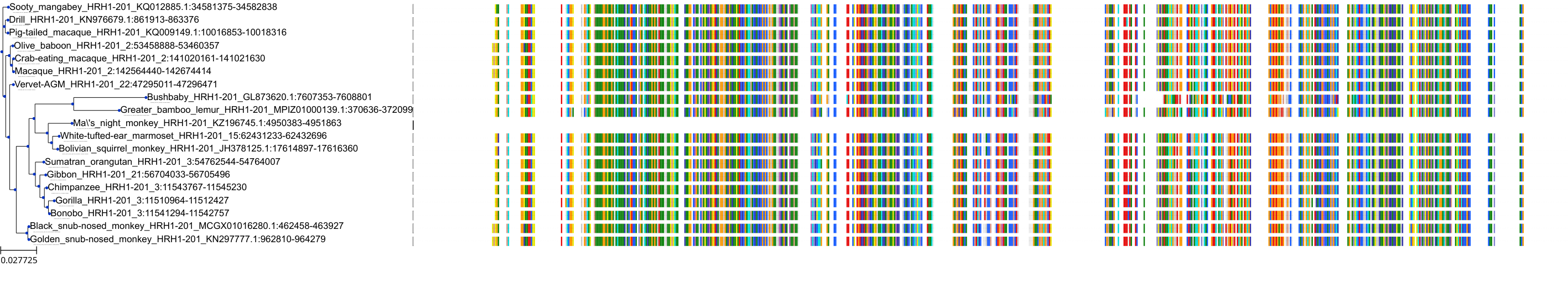
Target Conservation
|
Protein: Histamine H1 receptor Description: Histamine H1 receptor Organism : Homo sapiens P35367 ENSG00000196639 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 135954 |
| ChEMBL | CHEMBL1742423 |
| DrugBank | DB11591 |
| DrugCentral | 4353 |
| FDA SRS | PA1123N395 |
| Human Metabolome Database | HMDB0240232 |
| PubChem | 185460 |
| SureChEMBL | SCHEMBL991810 |
| ZINC | ZINC000003822702 |