| Trade Names | |
| Synonyms | |
| Status | |
| Molecule Category | Mixture |
| ATC | A10BK02 |
| UNII | 6S49DGR869 |
Structure
| InChI Key | XTNGUQKDFGDXSJ-ZXGKGEBGSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C24H27FO6S |
| Molecular Weight | 462.54 |
| AlogP | 2.97 |
| Hydrogen Bond Acceptor | 6.0 |
| Hydrogen Bond Donor | 4.0 |
| Number of Rotational Bond | 5.0 |
| Polar Surface Area | 90.15 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 3.0 |
| Heavy Atoms | 31.0 |
Pharmacology
| Mechanism of Action | Action | Reference |
|---|---|---|
| Sodium/glucose cotransporter 2 inhibitor | INHIBITOR | DailyMed |
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Transporter
Electrochemical transporter
SLC superfamily of solute carriers
SLC05 family of sodium-dependent glucose transporters
|
2.2-910 | 2.2-684 | - | - | - |
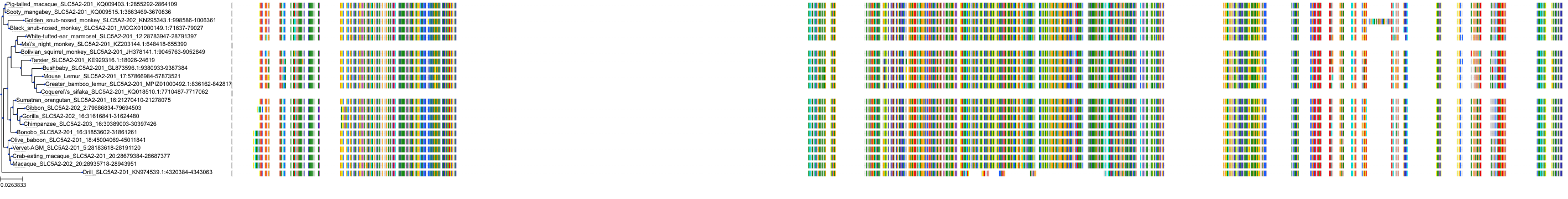
Target Conservation
|
Protein: Sodium/glucose cotransporter 2 Description: Sodium/glucose cotransporter 2 Organism : Homo sapiens P31639 ENSG00000140675 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 73274 |
| ChEMBL | CHEMBL2048484 |
| DrugBank | DB08907 |
| DrugCentral | 4758 |
| FDA SRS | 6S49DGR869 |
| Guide to Pharmacology | 4582 |
| SureChEMBL | SCHEMBL157162 |
| ZINC | ZINC000043207238 |
| ChEMBL | CHEMBL4594217 |
| FDA SRS | B2L6AES9XQ |
| SureChEMBL | SCHEMBL322059 |
 Homo sapiens
Homo sapiens