| Trade Names | |
| Synonyms | |
| Status | |
| Molecule Category | Free-form |
| ATC | S01EC01 |
| UNII | O3FX965V0I |
| EPA CompTox | DTXSID7022544 |
Structure
| InChI Key | BZKPWHYZMXOIDC-UHFFFAOYSA-N |
|---|---|
| Smiles | |
| InChI |
|
Physicochemical Descriptors
| Property Name | Value |
|---|---|
| Molecular Formula | C4H6N4O3S2 |
| Molecular Weight | 222.25 |
| AlogP | -0.86 |
| Hydrogen Bond Acceptor | 6.0 |
| Hydrogen Bond Donor | 2.0 |
| Number of Rotational Bond | 2.0 |
| Polar Surface Area | 115.04 |
| Molecular species | NEUTRAL |
| Aromatic Rings | 1.0 |
| Heavy Atoms | 13.0 |
Pharmacology
| Targets | EC50(nM) | IC50(nM) | Kd(nM) | Ki(nM) | Inhibition(%) | |
|---|---|---|---|---|---|---|
|
Enzyme
Hydrolase
|
- | - | - | - | 88 | |
|
Enzyme
Lyase
|
- | 2.58-999.8 | 1-840 | 0.012-900 | 13-100 | |
|
Enzyme
|
- | 2.58-999.8 | 1-840 | 0.012-900 | 13-100 | |
|
Transporter
Electrochemical transporter
SLC superfamily of solute carriers
SLC21/SLCO family of organic anion transporting polypeptides
|
- | - | - | - | 84.51-90.74 |
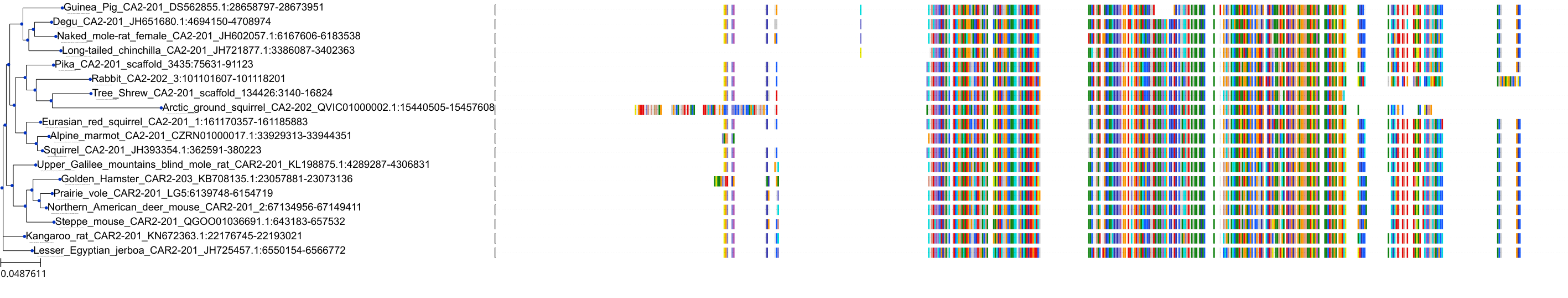
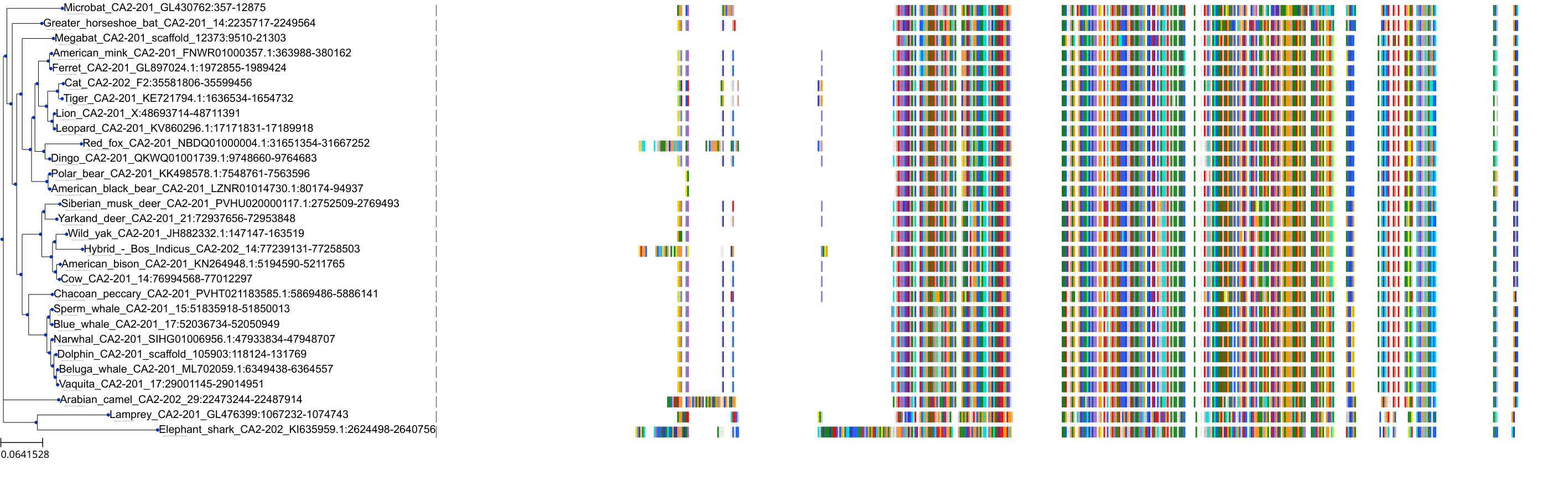
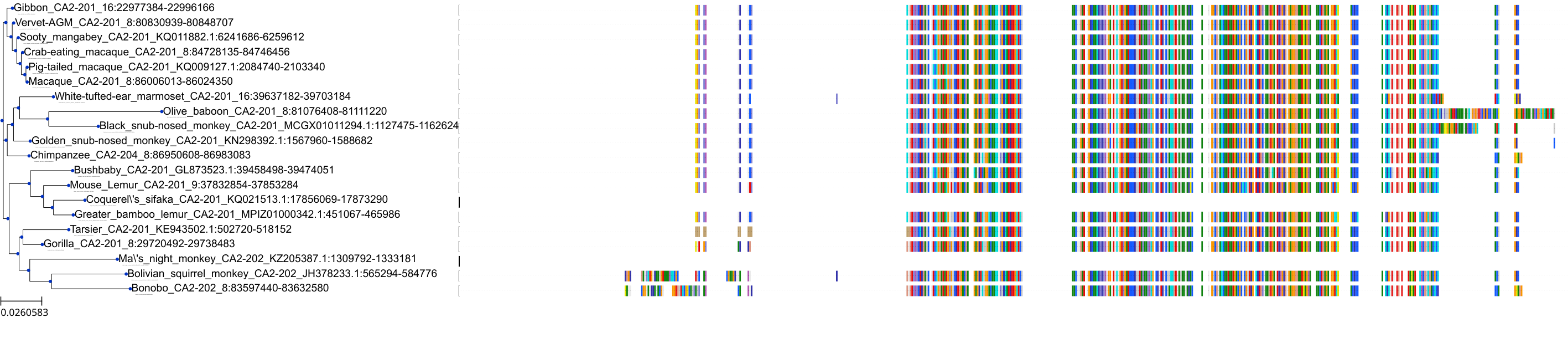
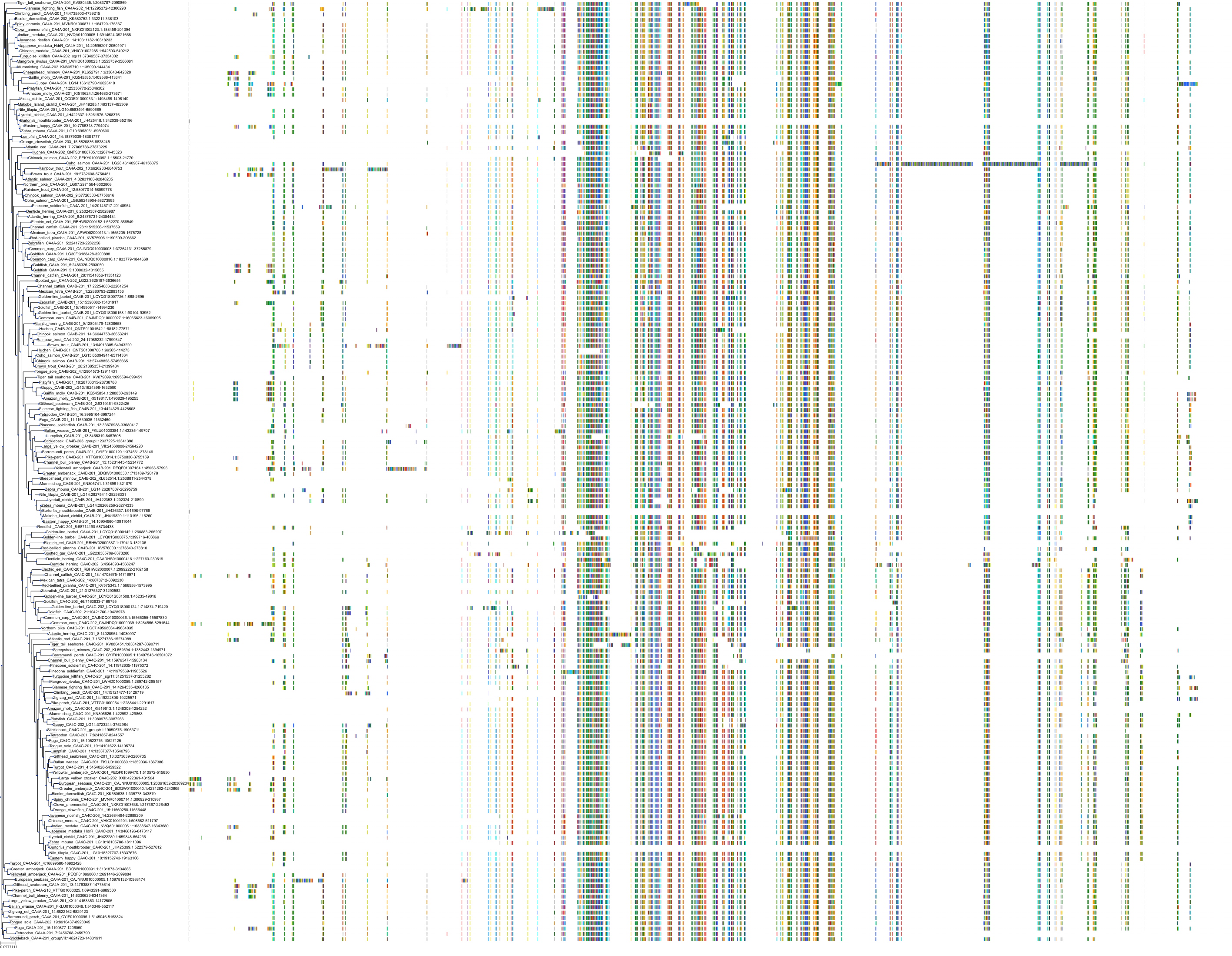
Target Conservation
|
Protein: Carbonic anhydrase XII Description: Carbonic anhydrase 12 Organism : Homo sapiens O43570 ENSG00000074410 |
||||
|
Protein: Carbonic anhydrase I Description: Carbonic anhydrase 1 Organism : Homo sapiens P00915 ENSG00000133742 |
||||
|
Protein: Carbonic anhydrase II Description: Carbonic anhydrase 2 Organism : Homo sapiens P00918 ENSG00000104267 |
||||
|
Protein: Carbonic anhydrase IV Description: Carbonic anhydrase 4 Organism : Homo sapiens P22748 ENSG00000167434 |
||||
Cross References
| Resources | Reference |
|---|---|
| ChEBI | 27690 |
| ChEMBL | CHEMBL20 |
| DrugBank | DB00819 |
| DrugCentral | 56 |
| FDA SRS | O3FX965V0I |
| Human Metabolome Database | HMDB0014957 |
| Guide to Pharmacology | 6792 |
| KEGG | C06805 |
| PDB | AZM |
| PharmGKB | PA448018 |
| PubChem | 1986 |
| SureChEMBL | SCHEMBL23219 |
| ZINC | ZINC000003813042 |
 Anopheles gambiae
Anopheles gambiae
 Aspergillus fumigatus
Aspergillus fumigatus
 Astrosclera willeyana
Astrosclera willeyana
 Bos taurus
Bos taurus
 Brucella suis
Brucella suis
 Burkholderia pseudomallei
Burkholderia pseudomallei
 Caenorhabditis elegans
Caenorhabditis elegans
 Candida albicans
Candida albicans
 Candida glabrata
Candida glabrata
 Chionodraco hamatus
Chionodraco hamatus
 Coleofasciculus chthonoplastes
Coleofasciculus chthonoplastes
 Colwellia psychrerythraea
Colwellia psychrerythraea
 Cricetulus griseus
Cricetulus griseus
 Cryptococcus neoformans
Cryptococcus neoformans
 Cryptococcus neoformans var. grubii
Cryptococcus neoformans var. grubii
 Drosophila melanogaster
Drosophila melanogaster
 Enterobacter sp.
Enterobacter sp.
 Filobasidiella neoformans
Filobasidiella neoformans
 Flaveria bidentis
Flaveria bidentis
 Francisella tularensis
Francisella tularensis
 Helicobacter pylori
Helicobacter pylori
 Homo sapiens
Homo sapiens
 Legionella pneumophila
Legionella pneumophila
 Legionella pneumophila subsp. pneumophila str. Philadelphia 1
Legionella pneumophila subsp. pneumophila str. Philadelphia 1
 Leishmania chagasi
Leishmania chagasi
 Leishmania donovani
Leishmania donovani
 Leishmania donovani chagasi
Leishmania donovani chagasi
 Leptonychotes weddellii
Leptonychotes weddellii
 Malassezia globosa
Malassezia globosa
 Methanosarcina thermophila
Methanosarcina thermophila
 Mus musculus
Mus musculus
 Mycobacterium tuberculosis
Mycobacterium tuberculosis
 Mycobacterium tuberculosis H37Rv
Mycobacterium tuberculosis H37Rv
 Nostoc commune
Nostoc commune
 Oryctolagus cuniculus
Oryctolagus cuniculus
 Plasmodium falciparum
Plasmodium falciparum
 Porphyromonas gingivalis
Porphyromonas gingivalis
 Pseudoalteromonas haloplanktis
Pseudoalteromonas haloplanktis
 Pseudomonas aeruginosa PAO1
Pseudomonas aeruginosa PAO1
 Saccharomyces cerevisiae
Saccharomyces cerevisiae
 Salmonella enterica subsp. enterica serovar Typhimurium
Salmonella enterica subsp. enterica serovar Typhimurium
 Schistosoma mansoni
Schistosoma mansoni
 Streptococcus mutans UA159
Streptococcus mutans UA159
 Streptococcus pneumoniae
Streptococcus pneumoniae
 Struthio camelus
Struthio camelus
 Stylophora pistillata
Stylophora pistillata
 Sulfurihydrogenibium azorense
Sulfurihydrogenibium azorense
 Sulfurihydrogenibium yellowstonense
Sulfurihydrogenibium yellowstonense
 Thalassiosira weissflogii
Thalassiosira weissflogii
 Thiomicrospira crunogena XCL-2
Thiomicrospira crunogena XCL-2
 Trypanosoma cruzi
Trypanosoma cruzi
 Vibrio cholerae
Vibrio cholerae